
Unlocking the complexity of proteins to fuel precision medicine (Part 2)

Tyler Ford
April 20, 2023
When the first draft of the human genome was completed in the early 2000s, many hoped the ensuing genomics revolution would usher in a new era in drug development. Researchers would use their understanding of the genome to create personalized “precision medicines” that target diseases at their root genetic causes and have a high likelihood of clinical success. However, this has not been possible for most diseases and only around 13% of all drugs entering clinical trials make it to market (Wong et al 2019). Billions of dollars still go into developing drugs that ultimately fail (Wouters et al 2020). Here we describe how proteomics can radically improve the drug development process.
Identifying potential drug targets
Even if we have some insights into the mechanisms of disease, it can be difficult to establish possible drug targets. For that, we need an empirical evaluation of protein abundance in diseased and healthy cells. Proteomics technologies can identify proteins with low abundance in healthy cells and high abundance in diseased cells. Proteomics can also tell us whether these proteins are at cellular locations that are easy to reach with drugs.
For example, with proteomics, researchers can identify proteins found at high levels on the surface of diseased cells but not healthy ones. These proteins may be easy to target with antibodies and other kinds of drugs.
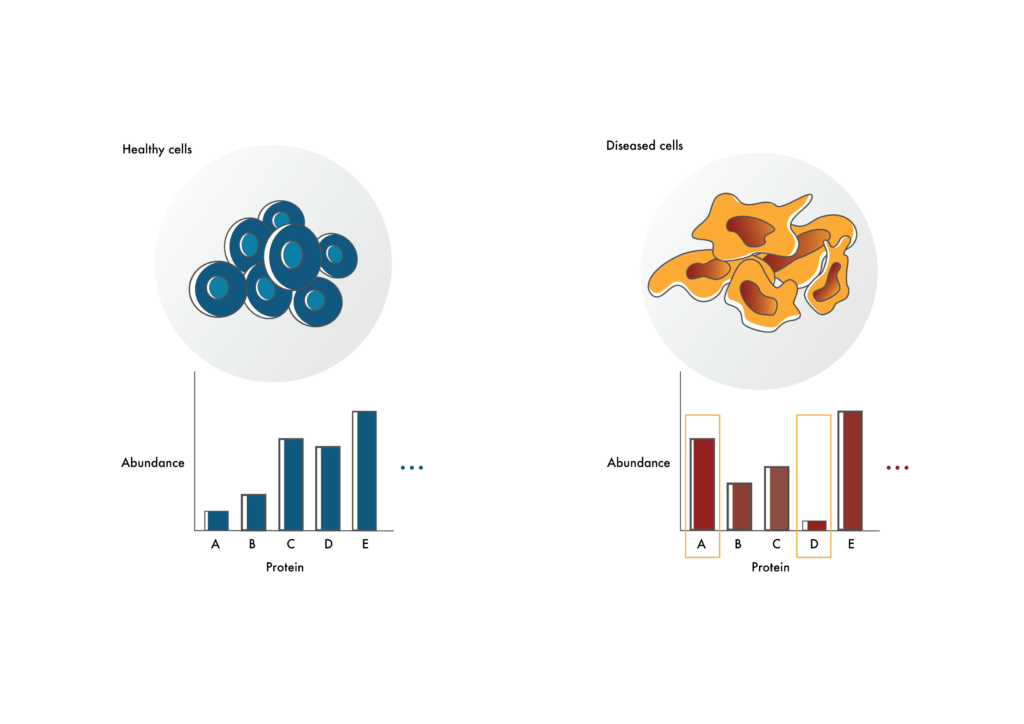
Identifying possible drug targets
Researchers can use proteomics to identify proteins that are at elevated or decreased levels in diseased cells compared to healthy cells (like proteins A and D). The proteins that are elevated in diseased cells may make good drug targets. Those with decreased levels may help explain disease mechanism.
Making effective drugs with few side effects
Drug developers cannot go straight from a list of protein abundances in healthy and diseased cells to the clinic. First, they must create drugs that interact with their protein targets in ways that actually impact disease. In addition, they must show that these drugs don’t cause side effects. Proteomics helps them do both.
Differences in the proteomes of healthy and diseased cells provide objective measures of disease. If a drug makes a diseased cell’s proteome look like that of a healthy cell, it is likely to be effective. Here the proteome, and potentially a select set of protein abundances, acts as a “biomarker.” That is, an objective, biological measure of drug effectiveness and disease severity.
Yet, even if a drug appears to mitigate disease, it may have unbearable side effects for patients. These can be caused by unanticipated drug-protein interactions. Proteomics can help drug developers avoid side effects by giving them a more holistic understanding of such drug-protein interactions.
Indeed, undesirable interactions like these manifest when drug developers focus too much on interactions between drugs and their intended targets. They may make drugs that bind really well to their targets. Yet, if they don’t know how a drug impacts other proteins, they won’t get a full understanding of its “mechanism of action.” The drug might bind to other “off-target” proteins causing “off-target toxicity.” It might even cause the target to interact with other cellular components in unexpected ways (Roberts 2018). If they choose a different target, they may be able to create drugs that interact with it more specifically. However, if they don’t start with a holistic understanding of drug-protein interactions, they won’t realize this is a problem.
Researchers don’t usually see the effects of unexpected drug-protein interactions until the drug enters the clinic. As a result, researchers create drugs that have small “therapeutic windows.” At a low dose, these drugs selectively interact with the appropriate targets and have positive effects. However, as the dose increases, unintended interactions begin and toxicity dominates.
We anticipate advances in analyzing and quantifying proteins will give researchers a more holistic understanding of drug-protein interactions. Before dosing patients and discovering all sorts of unexpected toxicity, drug developers can instead look for signs that the drug and/or interactions with its target cause unwanted changes to the proteome. They can eliminate potential targets before developing drugs against them and stop studies before entering expensive clinical trials.
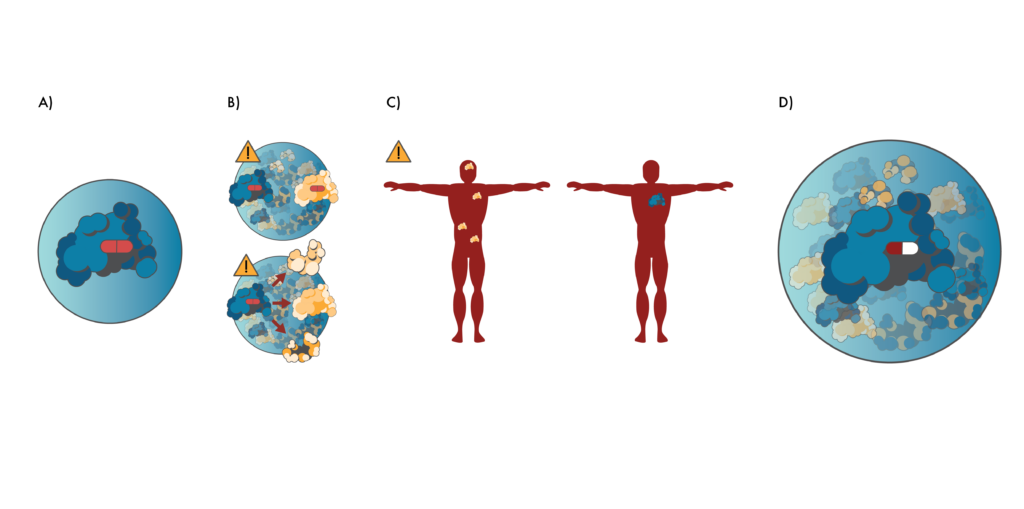
Preventing toxicity through proteomics.
A) Researchers often create drugs (red pill) that target proteins in isolation. B) However, this can lead to many unexpected interactions between the drug and other proteins (like the yellow protein above) once the drug enters cells. Alternatively, the drug may cause the target protein to interact with other proteins. These unintended interactions can lead to cellular malfunctions and toxicity. C) Drugs created this way may also target proteins that are expressed at many different places in the body (left, yellow protein). These may be bad drug targets if the proteins only cause disease associated with a particular part of the body. Better drug targets (right, blue protein) may only be expressed in the part of the body where they cause disease. D) Proteomics gives researchers a more holistic view of protein interactions during the drug development process. This enables them to create drugs (red/white pill) that are unlikely to have have “off-target” interactions and which target proteins that are only expressed at the disease site.
In the last post in this series, we’ll describe how the Nautilus Proteome Analysis Platform is designed to improve proteomics studies and may enhance drug development.
This blog post is an excerpt from the our white paper titled,“Proteomics and the development or precision medicines against cancer.” Download the full white paper here.
MORE ARTICLES

