
Traditional protein analysis methods – Western blotting, flow cytometry, and other methods using affinity reagents

Tyler Ford
October 27, 2022
The proteome is the full set of proteins within a biological sample like a cell, tissue, or organism. Proteomics is the study all of these proteins and their interactions through various protein analysis methods.
The particular proteomic make-up of any biological entity, including individual protein abundances and locations, determines how that entity works at the molecular level. Thus, mapping a cell’s proteome, also called broadscale proteomics, provides detailed information on biological activities and makes it possible to study the mechanisms behind these activities as well as disease states. Knowledge of the proteome lead to a broad range of discoveries and applications including everything from new therapeutics to new ways to protect plants from drought.
Given the essential importance of the proteome, scientists have been studying it for many years using various protein analysis methods like mass spectrometry, protein sequencing and more. In this “Traditional protein analysis” series, we cover how researchers typically study the proteins that make up the proteome.
These protein analysis methods are not necessarily “ omics” scale technologies, which generally aim to measure all or a large amount of a particular type of biological molecule in a sample. Nonetheless, these methods form the foundations upon which proteomic analysis methods have been built. In a future series, we will dive into the technical concepts behind emerging omics technologies, which aim to enable scientists to study the full proteome.
Comparing different protein analysis methods
In the graphics portraying the proteomic analysis tools below, we provide qualitative assessments of proteome coverage and ease-of-use for each. Technologies with low proteome coverage, like western blotting and flow cytometry, are generally used for targeted experiments analyzing a small number of proteins at once. Medium coverage protein analysis technologies like affinity arrays can look at 100’s to 1000’s of proteins in a single experiment. Truly comprehensive, high proteome coverage technologies can analyze tens of thousands of proteins at once.
Some protein analysis methods are easier to use than others. Low ease-of-use protein analysis tools generally require complicated, difficult, or customized sample preparation involving a lot of hands-on experimenter time, and their data may be difficult to analyze or require bioinformatics support. High ease-of-use protein analysis tools employ simple, standardized sample preparation, include more automation, and provide simple, data-rich outputs including protein abundance. Medium ease-of-use tools have some mix of these attributes.
In addition to assessing standard metrics like these, we also discuss some of the advantages and disadvantages of each protein analysis technique. Many protein profiling tools have specific pros and cons that make them ideal for certain proteomics applications, but not others.
Importantly, the technology assessments here should not be viewed as definitive. Rather, our goal is to help you think about how you can best leverage these technologies for your specific experimental goals.
Proteomics analysis methods: Antibodies and other affinity reagents
Antibodies are naturally produced proteins that enable organisms to protect themselves from foreign invaders. Antibodies generally bind to segments (epitopes) of other proteins. In doing so, they can prevent these target proteins from functioning. They can also recruit special immune cells to destroy the target protein or destroy the invading organism that produced the target protein.
For the purposes of protein analysis, scientists have different ways of creating antibodies that bind to proteins they would like to study. Researchers can use antibody-based protein analysis methods to determine the target protein’s presence, abundance, and location. Some of the antibody-based techniques researchers use to measure proteins include western blotting, affinity arrays, immunohistochemistry, and flow cytometry. All of these are covered below.
What is western blotting?
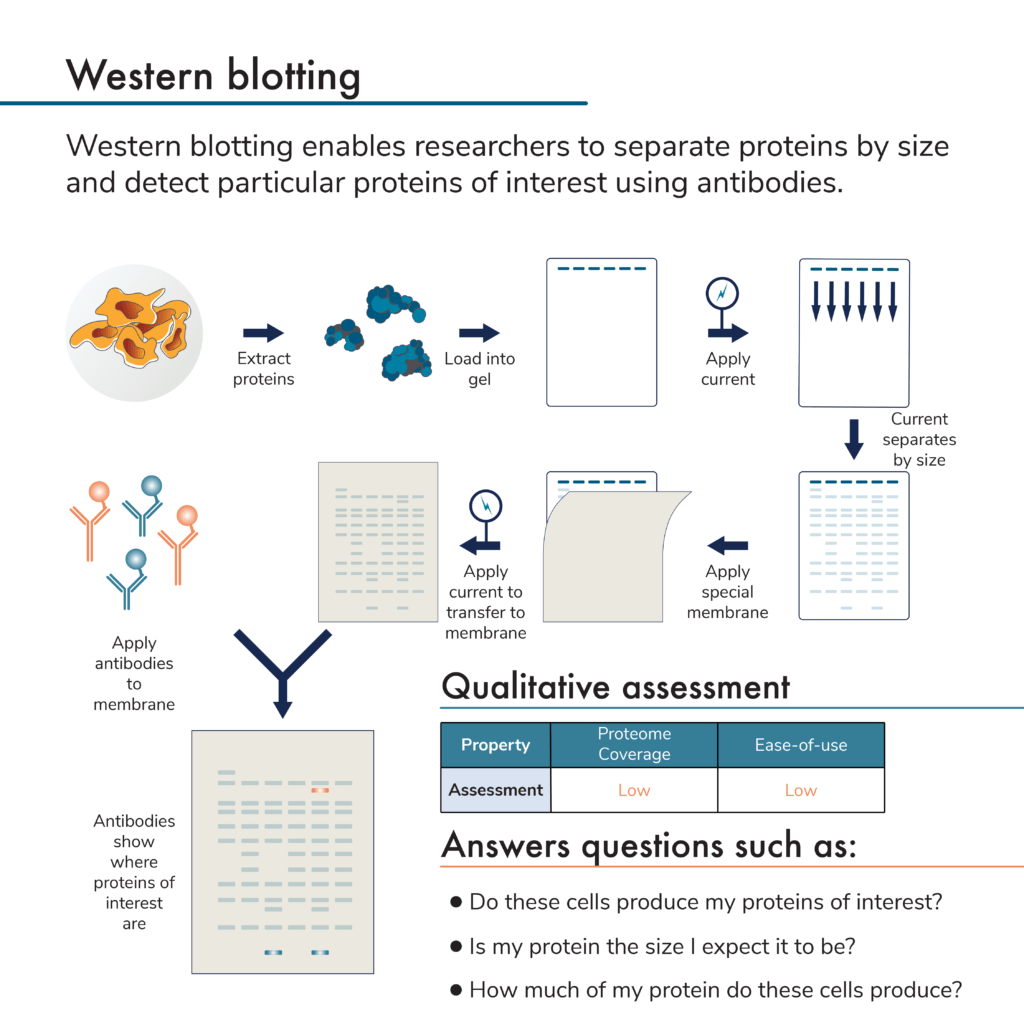
This is a low plex and low throughput protein analysis method. It involves first separating proteins by size using a porous gel. The proteins are then physically transferred from the gel to a membrane. Antibodies are applied and will bind to any target proteins stuck to the membrane. The presence of bound antibodies is then detected by imaging the membrane (directly detecting fluorescently-labeled antibodies, or using fluorescent secondary antibodies to bind to the target protein-antibody complex on the membrane).
By measuring the amount of antibodies sticking to the membrane, researchers can measure protein presence and relative abundance. Because the proteins are first separated by size, it is possible to determine the molecular weight of the target protein and multiple antibodies can be used on one membrane. Unfortunately, antibodies may also detect more than one protein due to a lack of perfect specificity. The need for an antibody specific to each target protein and the fact that antibodies may bind more than one target protein often limits how many target proteins a researcher can accurately study using this technique.
Advantages of western blotting
- Easily customizable – Many aspects of western blotting procedures, from extraction methods to visualization techniques, can be altered and optimized for a protein of interest.
- Low cost – Standard western blots do not require complicated equipment and allow researchers to perform protein analysis in just about any molecular biology lab with standard reagents. Higher throughput and automated systems can increase costs substantially.
- Can observe changes in protein size – Proteins undergo a variety of modifications, creating different proteoforms, that may result in changes to their size under different conditions. Because researchers separate proteins by size during the western blotting procedure, these changes can be measured (depending on the molecular weight resolution of the initial protein gel).
Disadvantages of western blotting
- Hands-on – While western blot procedures are straightforward, most steps must be performed manually. They often require extensive researcher time and attention and may be prone to operator error.
- Low proteome coverage – Western blots generally cannot be used to visualize more than a few proteins at once. Detection of any protein relies on having an antibody (either sourced commercially from vendors or developed in-house for new targets).
- Low throughput – Western blots generally accommodate tens of protein samples at most and take a few hours per target.
- Quantification is difficult – While western blots can be used to quantify proteins, this requires standards that can be difficult to make and use.
What are affinity arrays?
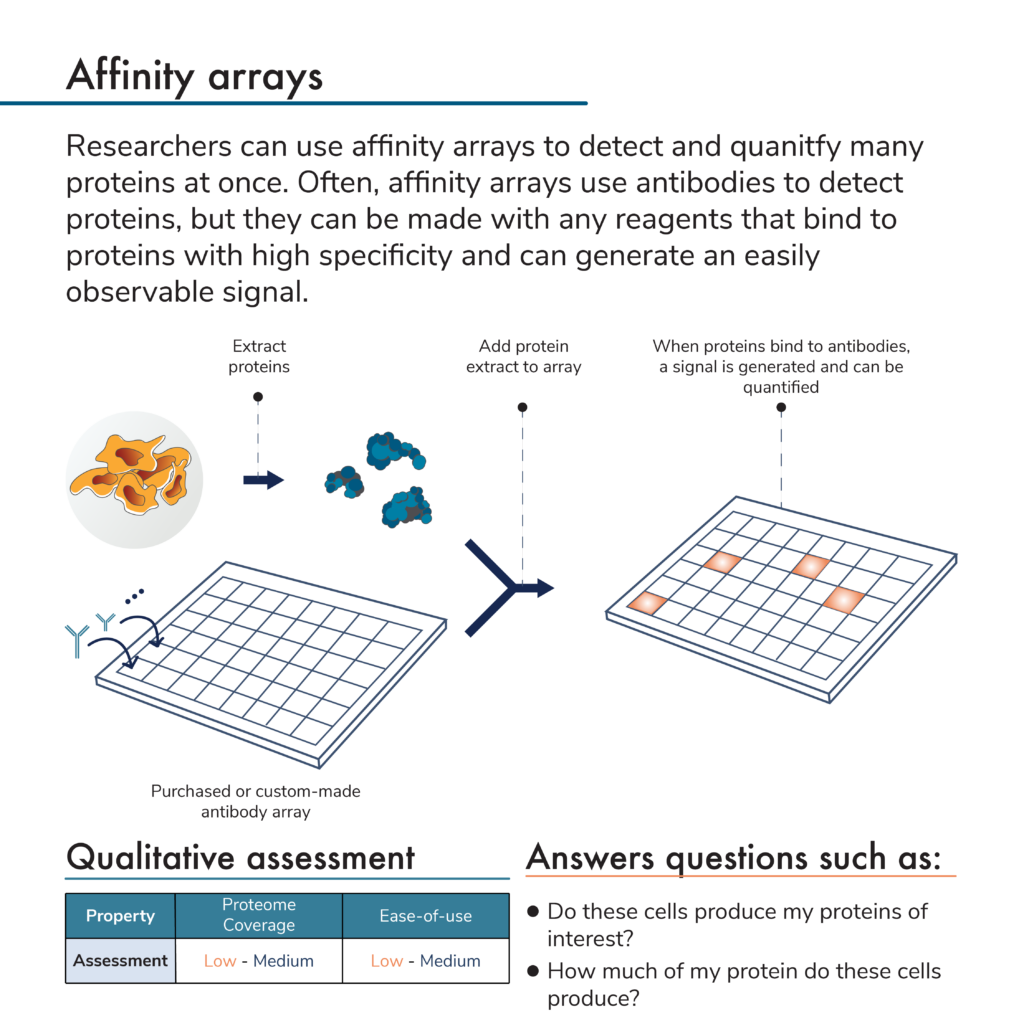
Rather than separate proteins and apply antibodies to them, researchers can do the opposite. That is, they can attach many different antibodies to specific locations on a solid surface to create an array of antibodies. An antibody microarray offers even more density, and higher coverage for protein analysis.
In this protein analysis method, researchers first flow a mixture of proteins from a sample over an antibody array. Later, they can add more reagents to the array to generate a signal indicating that a protein of interest is present. Because the signal will come from a known location with a known antibody that binds to a specific protein target, scientists can use these signals and their intensities to determine the identities and abundances of proteins in the original sample.
For some kinds of protein arrays, additional detection chemicals are not necessary. Instead, protein binding might change a physical or electrical property of the array, and this can be measured with specialized equipment for protein detection.
Advantages of affinity arrays for protein analysis
- Proteome coverage – Affinity arrays can detect more proteins at once compared to western blots. Nonetheless, there are many different kinds of arrays and some may only provide a very targeted assessment of a few proteins at once.
- Quantification – Depending on the detection technique, it may be easier to compare protein levels across standardized affinity arrays than western blots.
Disadvantages of affinity arrays for protein analysis
- Highly-validated antibodies are required – Proteins are not separated by size before detection with an affinity array. Thus, non-specific or off-target binding may be harder to differentiate from on-target binding on an affinity array compared to a western blot (which provides additional molecular weight information to support protein identification).
- Low ease-of-use – Affinity arrays can be highly standardized and working with them can be automated, but many arrays require customized sample prep, large amounts of hands-on time, and may be prone to operator error.
- Cost – Affinity arrays are much more expensive than the materials required for a western blot and the instruments required for imaging highly multiplexed arrays add even greater expense.
What is immunohistochemistry?
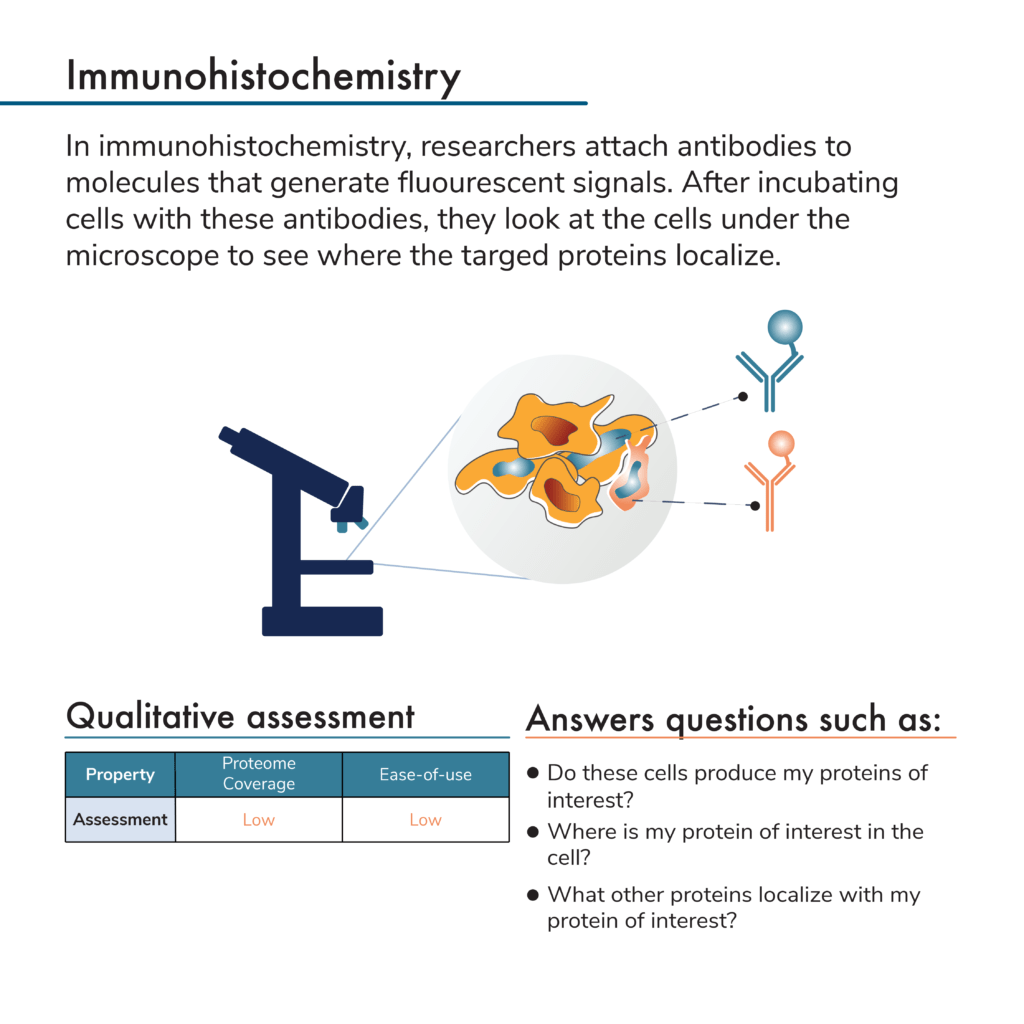
Neither of the above protein analysis methods provide any information about a protein’s location in a cell or in a tissue. To get that level of information with western blots or affinity arrays, researchers would first need to isolate proteins from particular cellular locations (i.e., from the nucleus or cytoplasm or from a particular section of tissue). Researchers can then analyze these extracts separately to determine what proteins they contain and where they came from. This adds significant work to the sample preparation process and is limited in resolution with respect to cellular location.
Antibodies can, however, be used to visualize and localize proteins in intact cells and tissues. One way to do this is with microscopy and a process known as immunohistochemistry. Here samples of interest are incubated with fluorescent antibodies. Often researchers use a variety of chemical treatments to fix their samples so proteins remain in their cellular locations throughout processing. After protein sample preparation, researchers can look at the cells or tissues under the microscope and see where the fluorescent signals from their antibodies are located. In this way, they can determine where their proteins of interest reside in the cells or tissues.
Advantages of immunohistochemistry for protein analysis
- Direct visualization of protein localization – While there are indirect ways to use western blots and affinity arrays to determine where proteins reside in cells or tissues, immunohistochemistry shows researchers exactly where their proteins are.
- Multi-protein labeling – Researchers can use antibodies that bind to different proteins and fluoresce at different wavelengths to determine where multiple proteins reside in the cell at once. They can do this with as many colors of fluorescence as their microscopes can resolve. This level of multiplexed analysis with multiple wavelengths will not match array based methods but can offer additional information about protein co-localization or interactions that would be difficult to determine with other technologies
Disadvantages of immunohistochemistry for protein analysis
- Proteome coverage – While multi-protein labeling is useful as noted above, researchers generally cannot look at more than a few proteins at once using immunohistochemistry techniques.
- Throughput – There are ways to automate the staining and visualization processes, but immunohistochemistry experiments are often done manually and require researchers to spend a large amount of time looking at their samples. Thus, they cannot analyze more than a few samples per day.
- Cost – Microscopes capable of detecting fluorescence, especially multiple fluorescent colors, can be very expensive.
- Low ease-of-use – Microscopes used for immunohistochemistry experiments often have high learning curves and analysis/quantification can be difficult.
What is flow cytometry?
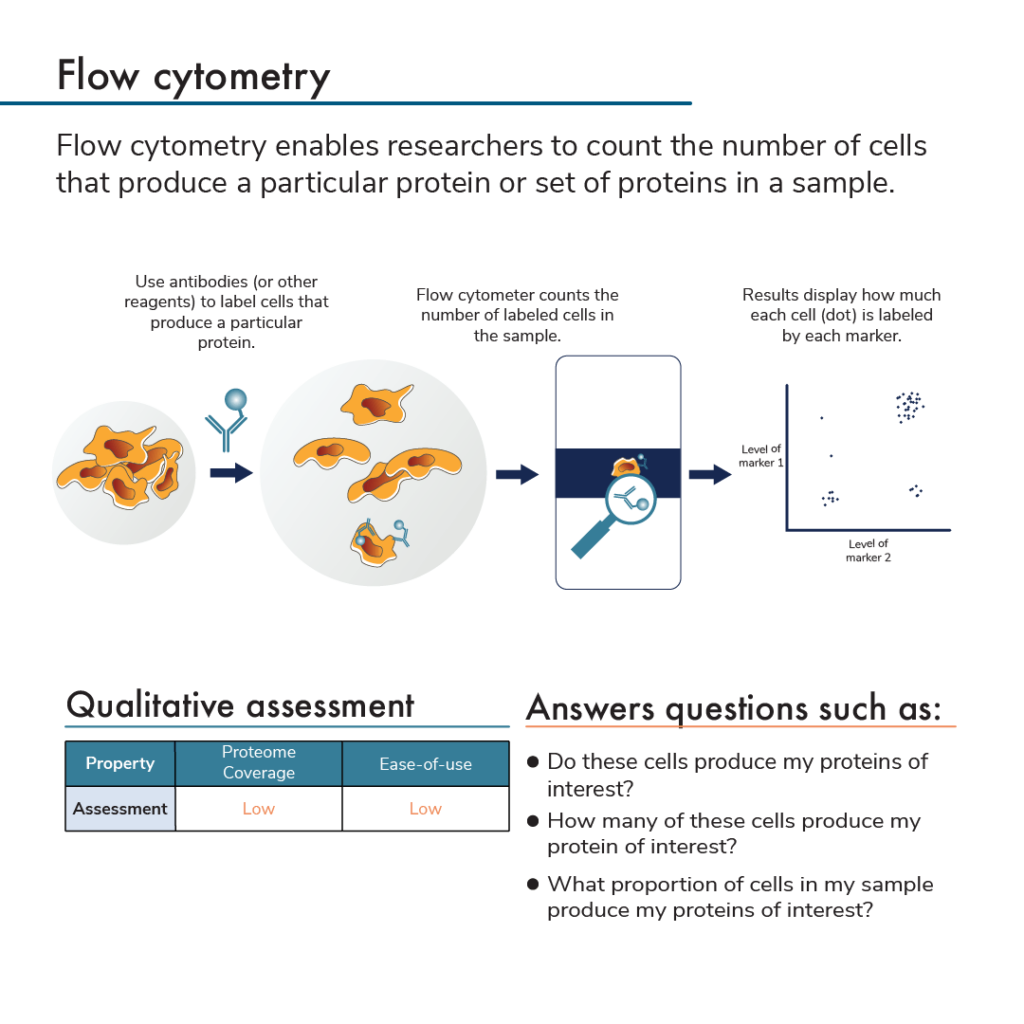
Protein analysis by flow cytometry uses labeled antibodies that bind to proteins expressed by subpopulations of cells. After incubating cells with antibodies, researchers insert the cells into a flow cytometer. This instrument analyzes individual cells and measures the amount of fluorescent label found on each cell. Flow cytometry also counts how many cells have a given set of labels. Specifically, the flow cytometer flows antibody-bound cells through small tubes that can only fit a single cell. A detector at the end of the tube identifies any labels found on the single cells as they pass through it. This can be done with as many colors/antibodies as a flow cytometer can distinguish.
Some flow cytometers can also be used for a protein analysis method called fluorescence activated cell sorting (FACS). When a FACS flow cytometer detects that a cell has a particular set of labels, it will shunt the cell into a particular pool. In this way, researchers can use flow cytometry analysis to specifically collect and study subsets of cells that produce specific groups of proteins.
Advantages of flow cytometry for protein analysis
- Multi-protein labeling – Similar to microscopy, researchers can label cells with antibodies that fluoresce different colors, count, and potentially sort cells that have specific combinations of labels. Using multi-protein labeling in a flow cytometer lets researchers specifically study cells that produce precise combinations of proteins.
- Protein quantitation – Counting the number of cells that express particular proteins with a flow cytometer can help researchers answer a variety of experimental questions. For instance, they might treat a pool of cells with a drug and determine if the drug specifically decreases the number of cells that produce a particular surface protein. Such a surface protein might be specific to cancer cells, for example.
- Cells can be analyzed after sorting – After discovering which of their cells of interest produce a particular protein, with FACS flow cytometry, researchers can continue to study these cells and their proteins using other.
Disadvantages of flow cytometry for protein analysis
- Cost – Flow cytometers and FACS instruments can be very expensive.
- Proteome coverage – With flow cytometry, researchers generally only analyze a few proteins at once.
- Low ease-of-use – Flow cytometers and FACS instruments can have a high learning curve and it is not always easy to decide what level of antibody fluorescence should separate one type of cell from another. This makes data analysis complicated.
Protein analysis is about more than just antibodies!
It is important to note that many of these protein analysis methods do not have to be performed with antibodies. There are multiple types of biomolecules that can bind to proteins including aptamers, antibody fragments, and even other proteins. As long as a set of molecules binds to particular proteins with high specificity, and binding can generate a detectable signal, scientists can use these molecules to study the proteome.
Learn about other protein analysis methods like protein sequencing and mass spectrometry in the next two posts in this series.
MORE ARTICLES
