Organisms must be dynamic to survive in ever-changing environments. At the molecular level, organisms can respond to their surroundings through the creation of proteoforms – the single-molecule variants of proteins found in biological systems. These are defined by their full sets of alterations whether they come from genetic variation, alternative splicing, post-translational modifications (PTMs), or any other source (Smith and Kelleher 2013). Such alterations work together to change protein structure and function, and by identifying proteoforms as opposed to individual alterations or PTMs, researchers can gain mechanistic insights into biological processes.
Read our preprint to discover how we use Iterative Mapping to analyze proteoforms.
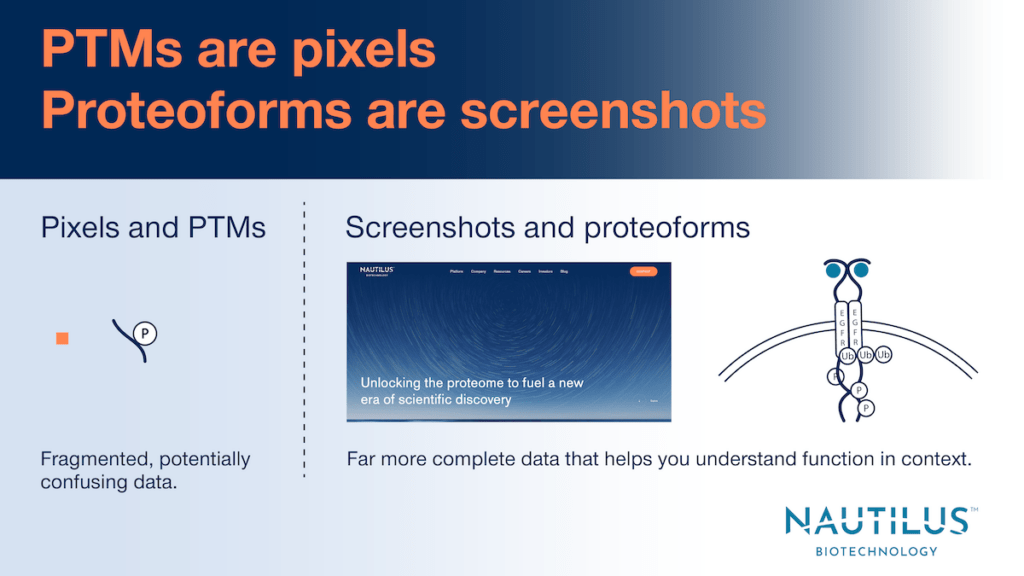
It is essential to study proteoforms as opposed to PTMs
PTMs and other isolated protein alterations can be misleading if studied in isolation. To understand why, consider a single phosphorylation event. To say this event activates or inhibits a protein may only be true for a certain isoform in the context of other PTMs. Thus, it is patterns of modification to single protein molecules that define their structures and functions. In other words, protein structure and function can be identified at the proteoform level but not the PTM level. Studying PTMs is like looking at isolated pixels on a monitor and attempting to discover what webpage is displayed. Studying proteoforms is like looking at screenshot – you get far more information that can lead to clear conclusions.
Examples of mechanistic insights derived from targeted proteoform analysis
Studies of EGFR showcase how proteoform analyses reveal information obscured in analyses of isolated PTMs. Upon EGF binding, EGFR autophosphorylates a subset of its tyrosine residues. Autophosphorylation of a particular EGFR tyrosine can be associated with either proliferative signaling or EGFR internalization and degradation. It all depends on whether a second tyrosine is also phosphorylated. When the first residue is phosphorylated in the absence of the second, the Grb2 protein is recruited and mediates proliferative signaling. When the second residue is also phosphorylated, Grb2 and this phosphosite cooperate to recruit a ubiquitin ligase that drives EGFR internalization and degradation (Sigismund et al. 2013).
These are just two of the many possible outcomes of EGFR signaling, but if one were to measure either of these phosphosites in isolation, even their associations with proliferative signaling and degradation would be difficult to interpret. They only become clear upon examining EGFR proteoforms. Furthermore, while it would be difficult to use either isolated phosphosite as a biomarker, EGFR proteoforms may effectively report on EGFR activity and downstream signaling.
The key is proteoforms can provide mechanistic understanding in instances where PTMs cannot. This is likely true for many, if not all proteins, and a few additional examples include:
- Histone modifications – one histone modification can enable another, and dynamic histone proteoforms cause epigenetic alterations to gene expression (Jain et al. 2023, Zhao et al. 2021).
- Enzyme kinetics – varying degrees of phosphorylation alter enzyme activity (Favelyukis et al. 2001).
- Neurodegenerative disease – Accumulated modifications to the tau protein correlate with increasing Alzheimer’s severity and may be involved in the mechanisms of tau aggregation (Wesseling et al. 2020).
The need for technologies that measure proteoforms and not just PTMs
Our current knowledge of proteoforms and their impacts often comes from low-throughput analyses involving protein purification and synthesis of defined proteoforms. In the last few decades, this work has advanced and researchers have begun to employ top-down proteomics to unambiguously associate proteoforms with specific phenotypes in a higher throughput manner (Melani et al. 2022). Nonetheless, techniques for measuring proteoforms as opposed to PTMs are rare, difficult to use, and unscalable. This must change if we hope to move beyond confusing catalogues of PTMs and ambiguous impacts on protein function.
Indeed, we must develop technologies that make proteoform analyses far more accessible so we can better understand how biological processes work and develop biomarkers that unambiguously report on their activity. With these technologies, we will additionally gain the incredible power to drug proteoforms with a precision that protein-targeted drugs do not have.
The NautilusTM Proteome Analysis Platform is uniquely suited to proteoform studies because single-molecule analysis lies at its technological core. The platform is designed to probe single protein molecules multiple times, discern their patterns of modification, and quantify proteoforms. Thus, we hope the platform will open the world of proteoforms to many more researchers and provide them with clear pictures of the active variants of proteins in biological systems. Learn more about proteoform analyses on the Nautilus Platform in this animation.
MORE ARTICLES