
Proteomics and the development of precision medicines against cancer (Part 1)

Tyler Ford
January 24, 2023
Cancer is a heterogenous mixture of diseases characterized by, among other things, the abnormal, uncontrolled growth of cells derived from otherwise healthy tissues (Hanahan 2022). Although cancer cells sometimes grow into balls of cells and stop there (so-called benign tumors), often they gain the ability to disperse throughout the body, seed the growth of other tumors, disrupt the function of a variety of organs, and ultimately kill patients. In this series of blog posts, we discuss how proteomics can drive the creation and use of “precision medicines” that target the processes enabling cancer growth and stop cancer in its tracks.
Want to read the full series now?
Targeting cancer with precision medicines
Cancer cells generally acquire the initial ability to multiply beyond their normal boundaries through mutations in genes that control cell growth. Such initial mutations set cells on a path to expand and divide rapidly, gain more mutations, attract blood vessels, and travel to other tissues (Hanahan and Weinberg 2011).
Recognizing the origin of cancer in such “driver” mutations, scientists have made many efforts to develop cancer treatments that target the proteins encoded in these mutated genes. Unfortunately, these efforts have only been successful for cancers with very strong driver mutations. For instance, a fusion of 2 genes can cause chronic myeloid leukemia (CML). The encoded fusion protein continuously signals for immune stem cells to grow and divide thereby leading to leukemia (Rumpold and Webersinke 2011). By targeting this protein with chemical (“small molecule”) drugs, so-called “precision medicines,” cancer growth is disrupted and most patients go into remission for years (Hochhaus et al 2017).
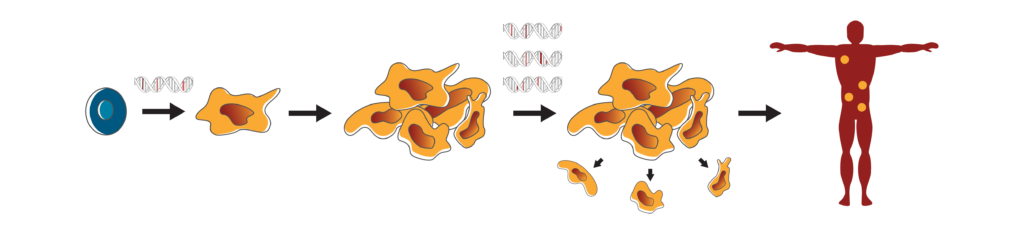
Driver mutations and cancer progression.
Driver mutations (red) cause healthy cells (blue) to transform into cancer cells (yellow) that grow and multiply beyond their normal, healthy boundaries. Although a single mutation (red) may be responsible for cancer development, there will likely be many other mutations (pink) that are inconsequential and it is hard to tell the difference between such mutations using genomic sequencing data alone. As cancer progresses, cancer cells acquire more mutations that enable them to continue to grow and spread throughout the body. Once again, the mutations that drive this process (red) come with inconsequential mutations (pink).
The need for proteome-powered precision medicines
Deciphering the means through which this fusion protein causes CML was by no means easy and it took decades to develop drugs against it. Nonetheless, many cancer-associated mutations cause disease in much less straightforward ways and are harder to develop precision medicines against. For example, cancer cells often have mutations comprising large inversions, duplications, deletions, insertions, or even translocations of long stretches of the 23 human chromosomes. One such translocation causes the gene fusion described above, but these chromosomal abnormalities can impact many genes at once and it is rarely obvious which of encoded proteins should be targeted with a drug.
Other times, there are many small changes to genes throughout a cancer cell’s genome. Many of these mutations will not have any impact on the encoded proteins and looking at DNA sequences alone is only the starting point for a scientist trying to understand their functional consequences.
Even if a scientist can identify a driver mutation in a specific gene, the encoded protein might not be easy to target with a therapeutic. For instance, scientists may not have chemical compounds that can bind to and affect the function of the mutated protein in any significant way. Thus, they may wish to look at the downstream impacts of this mutation to find proteins that are altered indirectly. These may be easier to target. For example, if a driver mutation in a protein known as a transcription factor causes it to enhance the production of growth-promoting proteins, scientists might want to target these growth-promoting proteins instead of the transcription factor. In this case, knowing the driver mutation alone would be insufficient for scientists to know what growth-promoting proteins to target. They would need to know how the production of other proteins changed as a result of the driver mutation. Proteins with increased abundance in cells containing the driver mutations might make good drug targets.
Overall, sequencing the DNA of cancer cells can give scientists clues as to the cause of a cancer and, in rare cases, may give them a precise protein to target therapeutically. However, in many cases, knowing information only about the static genome is neither enough to diagnose the particular type of cancer nor guide the best treatments.
Proteomics can complement DNA sequencing (genomics) and give researchers much more information about the mix of proteins found in cancer cells. Indeed, a cell’s “proteome” is its precise protein make-up including protein abundances, protein modifications, and protein distribution throughout the cell. Advanced proteomics technologies, like those being developed at Nautilus, aim to rapidly catalog the proteomes of cells grown in the lab or derived from patients. In the next two posts in this series, we discuss how advanced proteomics technologies improve cancer diagnostics and therapeutic outcomes.
Want to read the full series now? Download the “Proteomics and the development or precision medicines against cancer” white paper here.
View our animation see how next-generation proteomics can fuel cancer research.
MORE ARTICLES
