Proteins are fundamental to life on planet Earth. Yet, with current tools, we can only see a small fraction of the proteome. Ideally, one would be able to characterize and measure all the proteins in a given sample, its entire proteome. Analysis at this scale will provide extensive information about processes inside cells, tissues, and whole organisms, unlocking new biological discoveries and therapeutic targets. Proteomic analysis at this scale is on the horizon.
Proteomic analysis aims to examine all the proteins in a sample or a targeted subset of the proteins in a sample using a variety of techniques. Scientists have been conducting proteomic analyses for years, but proteomic analysis generally cannot capture the complete proteome of a sample.
Technology is holding back the field of proteomics, as is often the case in biological research. While tools like affinity-based antibody microarrays, mass spectrometry, and peptide sequencing have begun to open up the secrets of the proteome, they all have drawbacks. Right now, no technology delivers the ideal proteomic analysis platform.
For that, we need next-generation proteomics tools. These are techniques and platforms designed to greatly expand the scope of what’s possible in proteomics today, and deliver a complete proteomic analysis covering every protein in a sample.
In this post, we outline the elements underpinning an ideal proteomics platform and highlight how we’re designing Nautilus’ next-generation proteomics technology to achieve them.
View our animations to discover how our platform is designed to provide comprehensive proteome coverage and enable proteoform analysis:
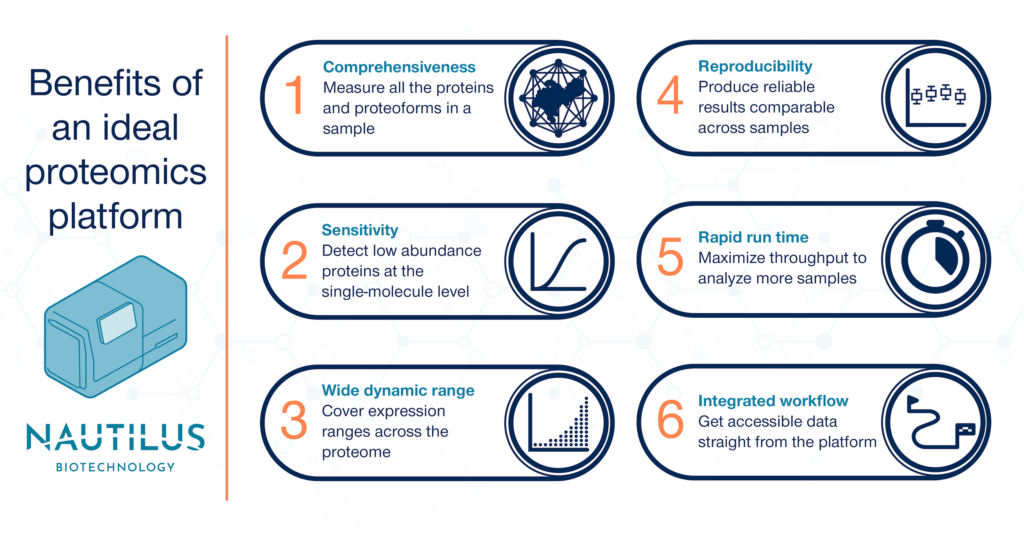
Elements of an ideal proteomics analysis platform
Comprehensiveness
With around 20,000 proteins in the human body, and countless variations of these proteins known as proteoforms, the human proteome encompasses millions of protein varieties. Furthermore, that’s just for our species — each species of plant, animal, fungus, virus, and bacteria has its own unique proteins. The variety of proteomes out there is simply massive. Nevertheless, to truly understand their complex functions in any biological entity, we need to be able to measure all proteins and proteoforms in a sample at once.
Thus, the first requirement of an ideal proteomic analysis platform is that it is comprehensive. To unlock the promise of proteomics, we need to see the vast diversity the proteome contains. Current techniques often see less than 10-30 percent of the proteome of any given sample — we need to do far better than that. Using hyperdense single-molecule arrays designed to capture and image 10 billion intact proteins at the single-molecule level, the Nautilus Proteome Analysis Platform has the potential to measure more than 95% of the human proteome.
Sensitivity
All proteins and proteoforms are not alike when it comes to abundance. Some are quite common, while others are rare in a given sample. But even these low-abundance proteins have roles to play. They may be biomarkers for diseases, targets for new drugs, or offer new insights into the mechanisms driving chronic conditions. With current proteomic analysis technologies, low-abundance proteins are often impossible to detect and remain hidden, hindering potential discoveries.
The second requirement of an ideal proteomic analysis platform is sensitivity. To encompass the scale of the proteome, we’ll need to see proteins that are expressed at very low levels compared to the proteome overall. With single-molecule measurements, the Nautilus Proteome Analysis Platform is designed to have the sensitivity to reveal these rare proteins.
Wide dynamic range
While we need the sensitivity to detect proteins present in small amounts, we’ll also need to identify and measure highly abundant proteins and everything in between simultaneously. Thus, we need a technology that can detect proteins across a wide range of expression.
Wide dynamic range is the third requirement for an ideal proteomic analysis platform. We need coverage ranging over many orders of magnitude in protein abundance to fully profile the proteome of any given sample. The Nautilus Proteome Analysis Platform is designed to measure 10 billion single, intact proteins per run. This equates to up to nine orders of magnitude of dynamic range, matching the scale of the proteome.
Check out this video for a visual dive into the dynamic range of the Nautilus Platform
Learn more about covering the wide dynamic range of the plasma proteome.
Reproducibility
Experimental results aren’t helpful if they change every time you run an experiment. Scaling experiments and translating discoveries requires that you trust your results, but this is not always possible with current proteomics platforms. For example, a significant drawback of mass spectrometry-based proteomics is that, while a mass spectrometer might measure 10-30 percent of the proteins in a sample, it may not measure that same 10-30 percent every time. That means the proteome, as seen by a mass spectrometer, will appear to change from sample to sample, requiring more samples to make confident conclusions.
The fourth requirement of an ideal proteomic analysis platform is reproducibility. Anyone interrogating the proteome of a sample wants to know that their results are accurate and trustworthy, every single time. The Nautilus platform is designed to deliver digital counts of protein abundance and provide reproducible results across samples.
Rapid run time
Another issue with current proteomic analysis technologies is that they’re slow. Low-throughput techniques slow down the pace of research and make some studies poorly suited for proteomic analysis. Unfortunately, many current proteomics technologies aren’t designed to allow for high-throughput experimentation.
Rapid run time is the fifth requirement of an ideal proteomic analysis platform. Faster proteomic analysis at scale means more discovery and less time wasted. Because it’s designed to conduct parallel analysis of multiple samples, the Nautilus Proteome Analysis Platform has the potential to deliver high sample and data throughput.
Integrated workflow
A technology’s workflow complexity is another factor that governs how long it takes to move from sample to data to actionable results. High complexity can also limit how many researchers can use a technique with confidence. Complicated, time-consuming workflows slow down science, increase the odds of mistakes, make onboarding new staff difficult, and make some technologies impossible to access broadly.
The sixth requirement of an ideal proteomic analysis platform is an integrated workflow. A simple, easy-to-understand process for sample preparation, protein identification/quantification, and data analysis speeds up proteomic research and opens opportunities to more labs. We’ve designed the Nautilus Platform with an integrated workflow in mind. The Platform is designed to generate easy-to-understand data including lists of proteins and their abundances expressed as counts. Our end goal is to make it easy to generate insights from your results.
Building a next-generation proteomics platform
Proteome analysis technologies that detect all proteins in a sample in a quick and reliable way will not only accelerate proteomics research but also democratize proteomics. By creating an easy-to-use next-generation proteomics platform that produces comprehensive and reliable results, we hope to make the proteome broadly accessible to scientists all over the world.
We can only guess at what this enabling technology might bring. Yet, given how essential proteins are to biology — and how little we still know about them — it’s likely that new proteomic insights will fundamentally change our understanding of the biological world and enable new therapeutic opportunities. With an ideal proteomics platform available to us, we have a better chance at curing diseases, fighting climate change, addressing food shortages, and more. Designing the right technologies is just the first step to enabling the proteomics revolution.
MORE ARTICLES