Applications of single-molecule proteomics – studying the tumor microenvironment

Tyler Ford
July 25, 2024
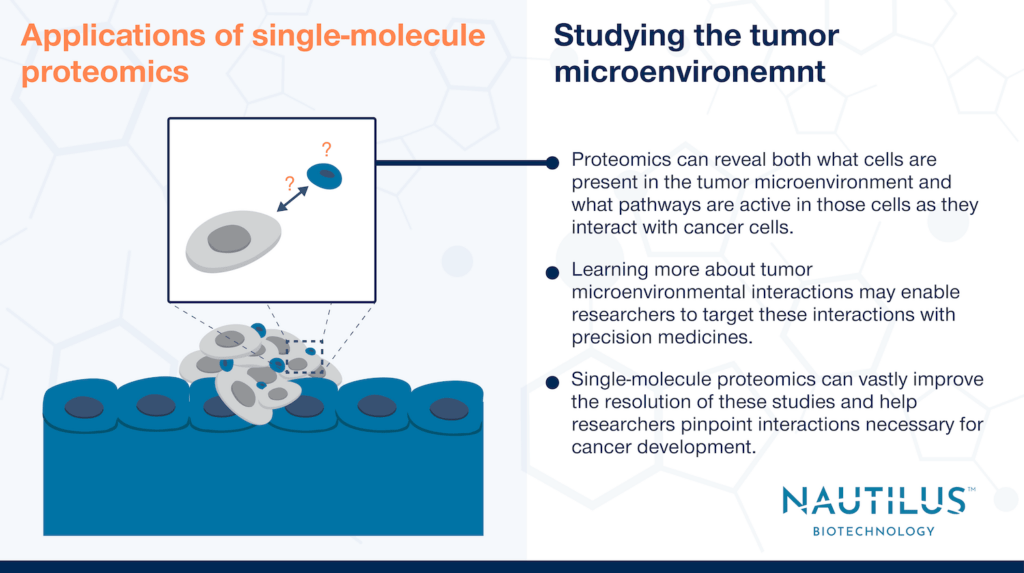
Nascent malignant cells and growing tumors do not live in isolation. They are surrounded by normal tissues with their own important functions. Throughout cancer growth and development, malignant cells must interact with these normal tissues in the “tumor microenvironment” for such varied activities as nutrient acquisition, migration, and immune defense. These microenvironmental interactions can be targeted to create more effective precision medicines against cancer.
Thus, while it is essential to study malignant cells to understand cancer progression, it is equally important to study how normal, adjacent cells interact with malignant cells. As with all things biological, studying the dynamic proteomes of these normal cells is perhaps the best way to understand how their functions and activities are impacted and even co-opted by cancer.
In this blog post, we cover fascinating work in which researchers leveraged mass spectrometry-based proteomics to learn about the mix of immune cells in the tumor microenvironment. Later, we explore some of the ways up-and-coming single-molecule proteomics technologies like the NautilusTM Proteome Analysis Platform may enhance similar studies in the future.
Deconvoluting the tumor microenvironment with proteomics
Petralia et al. 2024 leveraged multiomics data from over 1,000 tumors acquired and analyzed by the Clinical Proteomic Tumor Analysis Consortium to identify the cell types found and pathways active in tumors sampled from 10 cancer types. The authors only had data from bulk tumor samples so they used a bulk sample deconvolution algorithm called BayesDebulk to estimate the proportions of immune, stromal, and cancer cells within tumors. With the estimated cell types as well as protein abundance-based immune signatures, they could separate the tumors into seven immune subtypes.
Tumors from the seven different subtypes had important cellular and molecular differences with implications for treatment. Histological staining of a subset of tumors confirmed they had the expected immune cell percentages based on their subtype. The different subtypes also had disparate levels of signaling and metabolic pathway activation as assessed both by proteomics and phosphoproteomics. Furthermore, the authors applied their subtyping method to existing RNA-seq data from pre-treatment tumors and revealed that one subtype (CD8+/IFNG+) was more responsive to an immunotherapy.
Previous multiomic assessments of cancer had grouped tumors into immune subtypes based on pathway activity and transcriptomics, but this work employed the debulking algorithm to more concretely estimate the cell types present in each tumor. Doing so, the authors better defined the tumor microenvironment and delineated more subtypes. They could also point to pathways to modulate in some subtypes to potentially make them more responsive to immunotherapy.
Download our Proteomics and cancer eBook to learn more about ground-breaking cancer researcher leveraging proteomics
Applications of single-molecule proteomics in the study of the tumor microenvironment
Petralia et al. 2024 largely leveraged peptide-based mass spectrometry data for their analysis. They clearly learned a lot, but up-and-coming single-molecule proteomics techniques have the potential to expand this research. Single-molecule proteomics techniques have two main benefits:
- They are definitionally as sensitive as they can be
- If designed to detect full-length proteins, they enable researchers to comprehensively analyze proteoforms
For studies of the tumor microenvironment, sensitivity is crucial. Debulking algorithms can potentially be improved through more precise measurements of the proteins found in different cell types. These measurements inform the model parameters that define those cell types. When more precise sample measurements are then used as inputs for these refined models, researchers may get more accurate results. With the former, the algorithm may better attribute protein abundances to different cell types. With the latter, researchers can have more confidence in the protein abundance measurements and cell type estimates from their samples.
For researchers who forgo bulk analyses and focus on spatial proteomics or even single-cell proteomics, they’ll require the highest sensitivity possible to detect as much of the proteome as possible from their small samples. These techniques are bound to provide much higher resolution pictures of the cells present. Their development can potentially be accelerated through the inherent sensitivity of single-molecule proteomics.
Higher resolution views of proteoforms are also incredibly important. Comprehensive proteoform studies can identify the specific proteoforms present in various cell types and advance deconvolution efforts. They can also help researchers identify cancer specific biomarkers or drug targets. While Petralia et al. 2024 identified pathways active in their samples, it is specific proteoforms that often drive such pathways. Understanding what proteoforms are active in tumors will help researchers identify precise targets to manipulate in malignant cells specifically. This could result in more effective precision medicines.
High-resolution proteomic studies of the tumor microenvironment
Resolution is everything when studying the tumor microenvironment. We’re already learning much about the makeup of different cell types in tumors using current techniques. Yet, we need to learn more about the specific proteoforms and interactions that mediate outcomes to better target these interactions with precision medicines. Single-molecule proteomics can enable the development of new technologies that reveal the molecular details of these interactions and ultimately advance cancer care.
Watch this animation to learn how the Nautilus Platform is designed to quantify proteins at the single-molecule level
MORE ARTICLES