
Proximity proteomics for the identification of axonal proteomes – Applications of proteomics in neuroscience

Tyler Ford
August 22, 2023
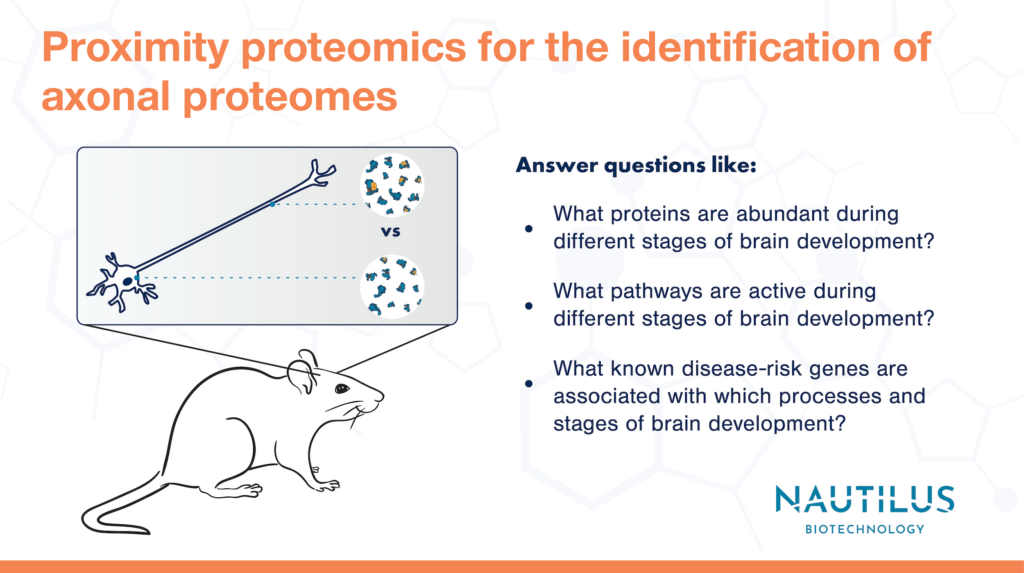
Many cells and tissues have complex structures that enable them to carry out their functions. For instance, neurons have long (sometimes incredibly long) extensions called axons that transmit electrochemical signals from one neuron to another. These wire-like connections have different protein compositions than neuronal cell bodies or “soma.”
Scientists endeavoring to understand how subcellular structures like axons work at the molecular level often look to the genes expressed within them. They use genomic tools to mutate genes and assess the impacts on cellular activities, but this does not provide direct insights into protein function. They also use transcriptomics and other RNA-based techniques to quantify differences in gene expression. However, special techniques are required to isolate RNA from axons specifically, and RNA expression levels do not always correlate with protein levels. This latter problem is compounded in studies of subcellular gene expression because proteins can be translated in one part of the cell and transported to another leading to a mismatch between local RNA and protein levels. For these reasons, otherwise powerful genomic and transcriptomic technologies cannot reveal what proteins are active in axons and driving function at a molecular, mechanistic level.
What is proximity proteomics?
Proximity proteomics is a technique in which researchers engineer cells to localize a labeling protein (often APEX) to a subcellular region of interest. Under the appropriate conditions, the APEX labeling protein causes biotin molecules to attach to any nearby proteins. Biotin-labeled proteins can be purified and later identified with a proteomic technique (usually mass spectrometry currently). Scientists can thereby use proximity proteomics to define subcellular proteomes.
This blog post highlights recent work by Vasin Dumrongprechachan et al. 2022. They use proximity proteomics to answer an important basic research question, “What proteins are enriched in axons?” Their findings are a great example of the power of proteomics and pave the way for studies investigating the roles of axonal proteins in a variety of behaviors and neurological diseases using next-generation proteomics technologies like the Nautilus Platform.
Watch our animation to see how next-generation proteomics can fuel neuroscience
Axonal proximity proteomics in a mouse model
To accomplish the identification of axonal proteins, these researchers created a mouse line expressing the APEX protein after cre-induced activation in a specific subset of neurons. Their APEX protein also had a nuclear export signal that effectively trafficked the protein away from the neuronal nucleus/soma to axons.
The researchers activated axonal APEX in brain tissues derived from this mouse line and did the same in control mice expressing APEX in the soma. Later, they extracted labeled proteins from the brain tissues at various developmental time points and identified the proteins via mass spectrometry. Proteins that were more abundant in the mice with axonal APEX were considered axon-enriched proteins.
Using bioinformatics techniques, the researchers clustered the axon-enriched proteins according to developmental expression patterns and thereby identified proteins and pathways important for axon maturation during different developmental time points.
With their mass spectrometry setup, the researchers could also identify phosphorylated proteins and use bioinformatic tools to determine which kinases and kinase pathways were active at the different stages of development. For example, the FYN kinase was particularly active during early postnatal development.
These meticulous efforts generated an extensive dataset mapping protein levels and activity to axonal development. Future researchers can mine this data to identify proteins important for neuronal processes occurring at each time point and even associate subsets of proteins with broader organismal behaviors or disorders.
Associating disease proteins with axonal development
As an example of how this data could be applied in disease research, the authors checked to see if there were any significant associations between known neurological disease-risk genes and the clusters of axonal protein expression. They found significant association between specific protein clusters and disease risk genes for autism spectrum disorders, bipolar disorders, epilepsy, and Alzheimer’s disease. Depending on which protein clusters/processes these risk genes were associated with, the researchers hypothesized functional roles for the risk genes.
For example, glutamate is a neurotransmitter involved in neuronal communication throughout the central nervous system. Past work has shown that mutations in glutamate receptor genes such as GRIN2B are associated with epilepsy. In this work, the glutamate receptor encoded in GRIN2B increased in abundance over time along with a cluster of other proteins, many of which were also epilepsy risk genes. In addition, the authors found this cluster was enriched for proteins in glutamate signaling. This reinforces the importance of glutamate signaling in epilepsy and implies that other epilepsy risk genes are linked to this essential process.
New proteomics tools for a better understanding of the brain and its many connections
We glossed over many of the details in the creation of the mouse lines used in this study. Their creation involved the use of CRISPR, Cre-lox, viral vectors, breeding, and more. These studies thus required a ton of genetic, molecular biology, biochemistry, and bioinformatics expertise. Top that all off with the need for extensive expertise in complex mass spectrometry workflows, and it’s obvious why many labs currently find it difficult to perform this kind of foundational proteomics work.
The data generated from this study alone can be mined to associate proteins with behaviors, neurological processes, diseases, and more for years to come. Nonetheless, to test the hypotheses generated by these associations, in-depth proteomics studies must be more accessible to researchers with diverse kinds of expertise.
We are developing the NautilusTM Proteome Analysis Platform with the goal of making comprehensive proteomic analyses like these more accessible to more researchers. Furthermore, we are designing our platform with higher sensitivity, dynamic range, and coverage than current proteomics technologies. With platforms like ours, neuroscientists will hopefully get even richer insights from studies like this one. If this “proteomics revolution” comes to fruition, there’s no telling what secrets of the brain scientists might uncover.
Check out our round up of proteomics and neuroscience posts.
MORE ARTICLES

