
Linking genes and proteins in neurological disease – Applications of proteomics in neuroscience

Tyler Ford
September 5, 2023
As our understanding of the neurological underpinnings of diseases affecting the brain grows, new therapies for previously intractable diseases like Alzheimer’s, Parkinson’s, and more may be within reach. As is often the case, these conditions also typically turn out to be far more complex than previously thought.
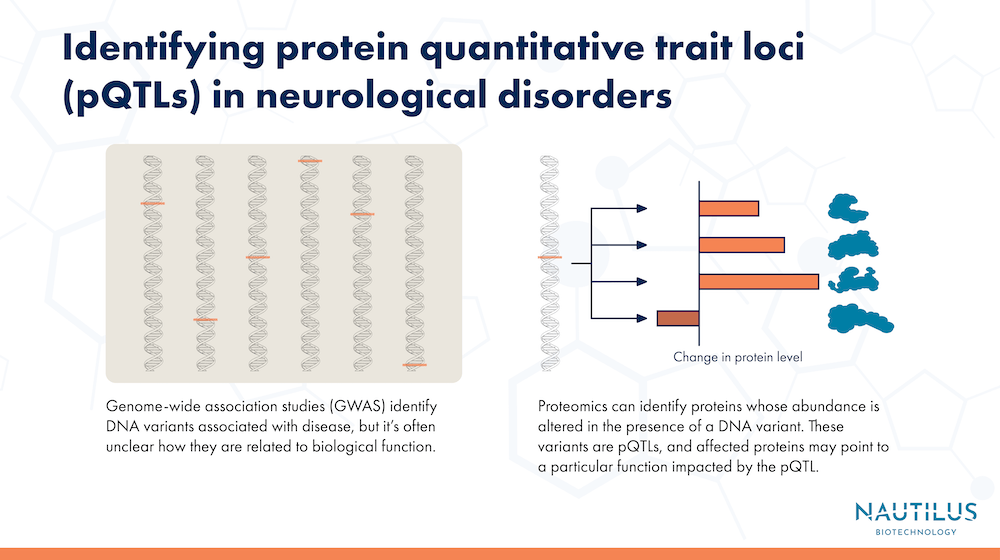
One example comes from genome-wide association studies, or GWAS. These large-scale studies use data from many different genomes to find correlations between an outcome of interest, like Alzheimer’s disease, and specific locations in the genome. One drawback of GWAS is that, while they’re good at pointing to genes that may be involved in a trait or disease, they often fail to show the underlying biological mechanisms involving those genes.
Another related issue is that, in some cases, genetic variants are tied to changes in protein expression, but not to changes in RNA, making it difficult to study the effects of these variants using studies of RNA expression.
Researchers can address this problem by turning to proteomics and directly studying the proteins involved in neurological disease. Indeed, next-generation proteomics technologies are making it possible to quickly and thoroughly interrogate the proteomes of diverse samples, opening the door to in-depth studies of the biological mechanisms behind disease.
In a recent multiomic analysis that combined genomic and proteomic techniques, Yang, et al were able to identify proteins across samples from different types of tissues affected by Alzheimer’s disease, stroke, and Parkinson’s disease. The research gives insights into the genes as well as the tissue-specific post-translational modifications key to neurological conditions.
The research underscores the power of proteomics to reveal the mechanisms behind biology in exquisite detail. It also highlights the complexities of translating genetic information into physical outcomes in organisms. By helping researchers get a better grip on how genes ultimately control protein levels in different areas of the body, this research could lay the groundwork for identifying new protein biomarkers for diseases, as well as novel drug targets.
Watch this animation to learn how next-generation proteomics technologies can fuel neuroscience
Linking genomic and proteomic analysis
To bridge the genome to proteome gap, the researchers looked for protein quantitative trait loci (pQTLs), regions of DNA that affect the abundance of specific proteins. They conducted a broadscale proteomic analysis of 1,744 samples from patients both with and without Alzheimer’s disease. The samples came from three different sources: brain tissue, cerebrospinal fluid, and blood plasma. All told, the researchers analyzed 1,305 different proteins, and performed GWAS to compare genetic variants from each patient with levels of the proteins in their tissues.
Identifying proteins associated with Alzheimer’s
With their proteomic analysis, the researchers found hundreds of pQTLs correlated with nearly as many proteins in the 3 different sample types. With further analysis, the researchers found 3 proteins in cerebrospinal fluid, 13 in plasma, and 7 in brain tissue that significantly affect the risk of Alzheimer’s disease. One such protein was CD33. This protein had been implicated in Alzheimer’s in other studies and affects how immune cells in the brain called microglia behave.
Their analysis also identified numerous individual proteins that were affected by multiple genes. They found 10 proteins that each had four genes acting on them, and one protein with five genes acting on it. Such multifactorial interactions highlight the complexity of protein regulation. Some of the identified genes could, for example, be governing when a particular protein is made, while others could be causing modifications to the protein after it’s made, changing how it behaves. Some of the genes could even be countering the actions of others, further muddying the waters.
Conversely, a number of genes also acted on more than one protein, an effect known as pleiotropy. One gene in particular, called apolipoprotein E, or APOE, was associated with changes in the levels of up to 13 different proteins. Versions of the APOE gene are known to be some of the strongest genetic risk factors for Alzheimer’s.
This data sheds more light on the complex genetic underpinnings of neurological conditions like Alzheimer’s disease. Identifying how specific proteins, and the genes that encode them, are involved in disease is a critical step toward developing drugs and other effective treatments for these conditions.
Next-generation proteomics for neurological diseases
In their paper, the researchers made important discoveries with a proteomic analysis covering over 1,300 proteins from patient samples. With millions of proteoforms in the human proteome, researchers will need better and more accessible proteomic analysis technologies to truly unlock the full proteome and expand on this exciting work.
Next-generation proteomics technologies like the Nautilus Proteome Analysis Platform are designed to enable scientists to analyze far more individual proteins at once, and open up proteomics to more labs. With a nanofabricated hyper-dense array, streamlined workflow, and unprecedented sensitivity and dynamic range, protein identification and quantification will hopefully happen more comprehensively and efficiently than ever before using the Nautilus Platform.
For multiomics studies digging into the links between genomics, proteomics, and other omics, accessibility and sensitivity may mean being able to see and understand far more connections than was previously possible. There are still genes that affect proteins in unknown ways, and proteins with functions that still aren’t understood because researchers haven’t been able to study them up close yet. The secrets to diseases like Alzheimer’s could lurk within that dark proteome — we’ll soon be able to bring them into the light.
Recent further research
Ali et al., 2025 used multiple proteomics platforms, samples, and kinds of analysis to identify potential Alzheimer’s biomarkers in cerebrospinal fluid. Their efforts enabled them to identify changes in proteins that had not been identified before and develop models for identifying cases of Alzheimer’s disease. Their final model had performance comparable to standard measures of CSF amyloid beta and phosphorylated tau. These new models also had higher specificity for Alzheimer’s when compared to previous proteomics models. Finally, their extensive datasets enabled them to track changes in the proteome across clinical stages of Alzheimer’s disease and identify changes in biological processes that may be important to disease progression.
Check out our round up of proteomics and neuroscience posts.
MORE ARTICLES

