
Applications of proteomics in neuroscience – Identifying disrupted pathways in dementia

Tyler Ford
August 15, 2023
When studying complex neurodegenerative diseases, it can be difficult to sift through mounds of data to determine how underlying biological processes are disrupted. More than that, it can be very hard to distinguish biological perturbations causing a disease from symptoms that occur during disease progression.
One way to get around this issue is to look for salient signals that appear in multiple types of data and see how these signals change across the course of a disease. Signals consistently appearing early on, and only in one specific disease, are more likely to be associated with the cause of the disease.
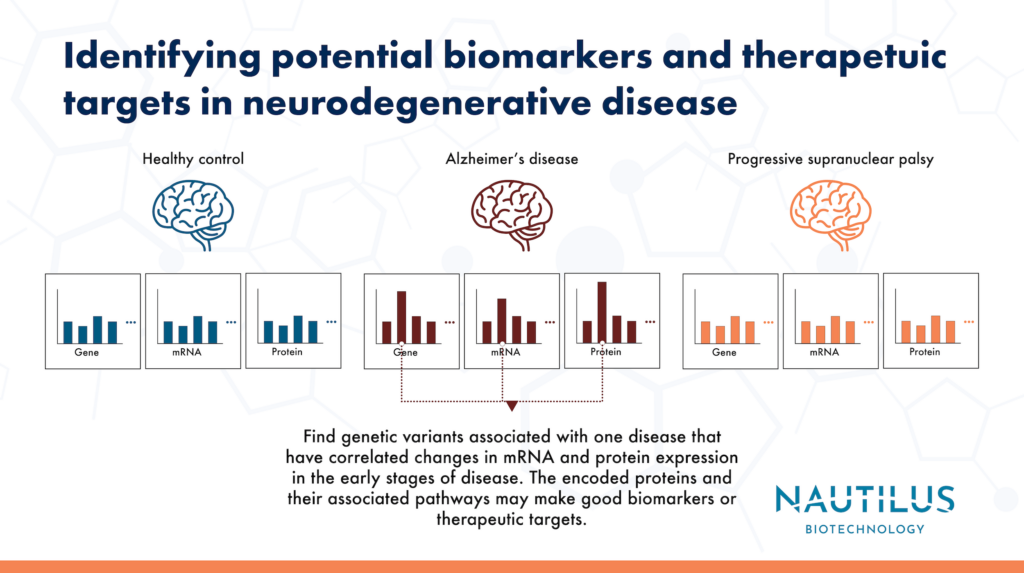
Swarup and Chang, et al use this strategy in their paper titled, “Identification of Conserved Proteomic Networks in Neurodegenerative Dementia.” Here, they leverage a multiomics approach to identify changes in proteomes, transcriptomes, and genomes of post-mortem brain samples from patients with various types of neurodegenerative disease. Concordant signals across various omics methodologies and samples point these researchers toward possible causes of dementia. Their results highlight potential biomarkers and therapeutic targets for neurodegenerative diseases.
Comparing proteomic changes across different neurodegenerative diseases
To understand how different diseases varied at the protein level, these authors analyzed the proteomes of post-mortem brain tissues from people with:
- Alzheimer’s disease
- Asymptomatic Alzheimer’s disease
- Progressive supranuclear palsy (PSP)
- Frontotemporal dementia
- Parkinson’s disease
- No disease (controls)
They found that particular protein modules associated with certain cell types like astrocytes, and functions like synaptic processes were either up-regulated or down-regulated in Alzheimer’s disease and these trends progressed along with disease severity. Looking at the other diseases, only those with dementia and not neurodegenerative diseases more broadly, recapitulated the trends found in Alzheimer’s disease. These findings implicate the identified protein modules in the specific progression of dementia.
In contrast to the protein modules perturbed early on in Alzheimer’s disease, one protein module perturbed in late Alzheimer’s was similarly affected across all the neurodegenerative diseases studied and not just those with dementia. This was the mitochondrial protein module. The authors postulate that cellular energetics are unsettled during the progression of many neurodegenerative diseases, and such disruptions may manifest late in disease as a consequence of many possible underlying molecular causes.
Watch our animation to see how next-generation proteomics can fuel neuroscience
Discordance between RNA and protein expression in neurodegenerative disease
Looking at proteomic and transcriptomic differences between Alzheimer’s disease and PSP (another type of dementia), the authors noted that, while changes in the transcriptome were often in the same direction as the proteome, they only accounted for ~50% of the variability at the protein level. This is a consistent finding across studies that assess concordance between RNA expression and protein expression (See Buccitelli and Selbach 2020 for a great review on the topic).
While incomplete concordance between RNA and protein expression can be a problem of measurement techniques, it is also an important characteristic of biological systems. There are a variety of cellular regulatory mechanisms that alter protein levels without necessarily impacting mRNA abundance. These can include processes like miRNA-based translational repression and enhanced protein degradation. Indeed, investigating the causes for discordance between mRNA and protein levels can reveal important regulatory mechanisms active in cells.
Nonetheless, in this work, there were distinct overlaps between the important players in the identified proteomic and transcriptomic modules. These may make high priority biomarkers or drug targets given their consistent association with disease across data types.
Even with these findings, the authors highlight that there were many genes whose protein expression correlated poorly with mRNA expression. The authors point to proteins from the electron transport chain and MAPK proteomic modules as prominent examples. Indeed, protein expression was even negatively correlated with mRNA expression for the electron transport chain module. This underscores the incredible importance of posttranscriptional and posttranslational regulation in modulating biological activity. It also calls attention to the need for proteomics to identify markers of biological function and dysfunction.
Combing genomics, transcriptomics, and proteomics to find disease-specific genes
These authors also checked to see if the modules with correlated changes in transcripts and proteins were enriched for genes associated with Alzheimer’s or PSP through Genome Wide Association Studies (GWAS). They found significant enrichment for GWAS-identified genes in modules associated with each disease and for early disease modules in Alzheimer’s in particular. In addition, they found that the enriched modules were not conserved across the two diseases. These results suggest that the GWAS-identified genes may be disease-specific or causal.
Of course, biology is always complicated. It is possible that the GWAS identified genes do impact late Alzheimer’s disease proteomic modules through trans effects. Even if a DNA variant is not directly in or near a particular gene, it may still impact that gene through effects on processes like transcription, translation, or mRNA and protein degradation. Nonetheless, these authors argue that concordance across the genome, transcriptome, and proteome further solidifies the identified genes and their products as potential biomarkers or therapeutic targets.
Enabling stronger comparisons across the omes with next-generation proteomics
The results from this study are particularly powerful because they are derived from multiple analyses across multiple sample types. Unfortunately, such studies are not easy to perform today because proteomic experiments at this scale are often cumbersome and do not cover the proteome in the comprehensive way that transcriptomics and genomics do. We are working tirelessly to ensure that accessible next-generation proteomics platforms like the Nautilus Proteome Analysis Platform will enable many more researchers to analyze comprehensive proteomic data along with genomic and transcriptomic data to identify potential biomarkers and therapeutics with high confidence. The insights obtained as a result will hopefully lead to care strategies and treatments that vastly improve the lives of those with neurodegenerative diseases.
Check out our round up of proteomics and neuroscience posts.
MORE ARTICLES

