Applications of proteomics in cancer – Proteomics reveals inner workings of head and neck squamous cell carcinoma (HNSCC)

Tyler Ford
November 17, 2023
Head and neck squamous cell carcinoma (HNSCC) is the sixth-most-common epithelial cancer, and it accounts for around four percent of all cancers in the United States. It includes tumors in the mouth, nose, larynx, throat, and tongue. One common risk factor for HNSCC is infection with the human papillomavirus (HPV); for this reason, HNSCC is broadly divided into HPV-positive and HPV-negative subtypes. The HPV-negative subtype typically has a much poorer prognosis, making it a high priority for new treatments.
Even within the HPV-negative classification, there can be significant differences in HNSCC cancer types involving different mutations, pathways, and more. A better understanding of HNSCC will include not only the genes that predispose people to this cancer, but the transcriptomic and proteomic factors salient to each subtype, allowing much more precise treatments.
In a 2021 paper in Cancer Cell, Huang et al. used a multiomic analysis to study HPV-negative HNSCC patient samples. Their analysis uncovered a wealth of information that could make HNSCC treatments more effective. Some of their high-level findings include:
- The connections between DNA alterations, mRNA expression, protein expression, and pathway activity in HNSCC are not obvious and require multiomic analyses to understand.
- Protein levels and data on phosphorylation status can identify potential pathways to target with precision medicines, including existing immunotherapy drugs.
- Combining multiomic data enables researchers to separate cancers into robust subgroups with actionable biomarkers.
HNSCC is a cancer with diverse drivers and biological mechanisms that make it difficult to study and treat. The multiomic research discussed here significantly advances our understanding of how various pathways are altered in this disease and could lead to new precision medicines for patients.
Check out this animation to see how next-generation proteomics can fuel cancer research
Proteomics applications in HNSCC
To better understand HPV-negative HNSCC, Huang et al. 2021 conducted a multiomic analysis encompassing genomics, epigenetics, proteomics, transcriptomics, and phosphoproteomics of 108 samples taken from patients with the cancer. Using mass-spectrometry-based proteomics, they compared the proteomes of tumor cells to the proteomes of matched samples from healthy tissues nearby. This discovery proteomics analysis revealed 3,355 proteins that were significantly increased and 3,163 proteins that were significantly decreased in tumors compared to healthy tissues.
Biomarkers indicative of HNSCC could help researchers better diagnose the disease, identify pathways to target therapeutically, and update prognoses. Looking at the proteins that were increased the most in tumors, the team uncovered 22 potential protein biomarkers for HNSCC, including seven currently targeted by FDA-approved drugs. There were also 162 proteins associated with progression-free survival that could be useful prognostic biomarkers.
Linking genomics and proteomics also helped reveal new insights into the mechanisms behind HNSCC. For example, the researchers looked at two common types of genetic alteration in HNSCC:
- Truncations to the FAT1 gene
- Amplification of the 11q13.3 region
They found that both types of mutation decrease actin protein levels with variable affects on actin mRNA. Amplification of 11.q13.13 was additionally associated with increased phosphorylation of an actin binding protein. Together, these results indicate that disrupted actin dynamics are an important component of HNSCC.
Their multiomic analysis also provided valuable insights into the regulation of the cell cycle in HNSCC through the cyclin D-CDK4/6-Rb pathway. It is often assumed that alterations to upstream genes CDKN2A or CCND1will ultimately change the phosphorylation status of the RB tumor-suppressor, and thereby impact the cell cycle. However, the researchers found aberrations in these genes did not always impact RB. Thus, one key conclusion is that RB protein status, and not just gene expression, is an effective and necessary indicator for CDK4/6-dependent cell-cycle activity. Only by looking at this pathway through a multiomic lens could the researchers accurately connect changes at the gene level to the output of the pathway.
Multiomic insights into HNSCC treatment
Huang et al. 2021 also uncovered new insights into why leading treatments for HNSCC fail in some patients. Mutated forms of the EGFR protein can drive runaway cell proliferation and are found in many kinds of cancer. Drugs based on monoclonal antibodies can inhibit EGFR, potentially helping control tumor growth, but the antibodies don’t work in every patient. From their proteogenomic analysis, Huang et al. 2021 saw that EGFR ligand abundance is predictive of successful monoclonal antibody therapy in HNSCC patents. Levels of EGFR gene amplification or overexpression on the other hand, were less predictive, a finding that may lead to better approaches to treating the cancer.
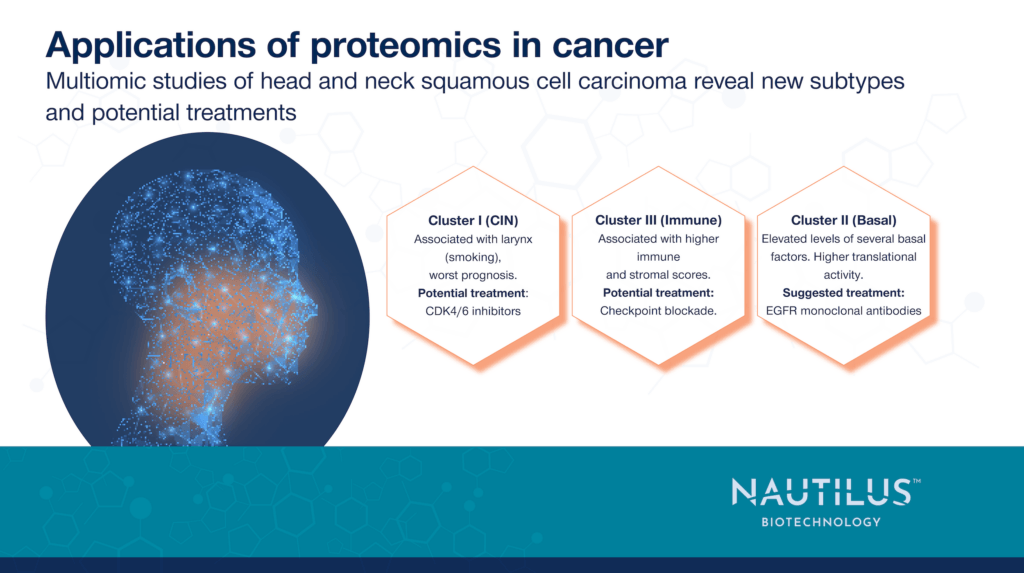
The team also identified three subtypes of HNSCC based on proteomic, transcriptomic, and other data, each characterized by the overexpression of different genes, and potential treatment options.
- Chromosome instability (CIN): characterized by mutations to the CCND1 and CDKN2A genes as well as high CDK4/6 activity. Cyclin-dependent kinase 4 and 6 (CDK4 and CDK6) inhibitors could be more effective in this group.
- Basal: characterized by elevated EGFR ligand expression and high EGFR pathway activity. Monoclonal antibodies targeting EGFR could be more effective in this group.
- Immune: characterized by elevated expression of immune checkpoint proteins. Immunotherapy drugs targeting checkpoints may be more effective in this group.
Next-generation proteomics tools for cancer research
By integrating proteomic insights with those from genomics and other disciplines in a multiomic framework, Huang et al. 2021 were able to glean insights into the fundamental biology of HNSCC, as well as identify potential biomarkers and avenues for targeted therapeutics. For instance, proteomics helped identify nuances in the way the EGFR protein behaves in different cancer patients, with clear clinical relevance for monoclonal antibody treatments.
Proteomic insights can be invaluable for studying disease biology, and, as the proteomics revolution progresses, the power of the proteome will only continue to grow. Including proteomics in a multiomic analysis is a powerful way to advance discovery and increase our knowledge of the molecular mechanisms behind various diseases and their subtypes.
Next-generation proteomics tools like the Nautilus Proteome Analysis Platform aim to make such analyses routine for researchers studying a wide variety of topics and reveal the vast potential of personalized treatments currently hidden in the depths of the proteome. We are designing our platform to make more studies like this one accessible to more labs and thereby spur the creation of more effective treatments for a wide variety of cancers.
MORE ARTICLES