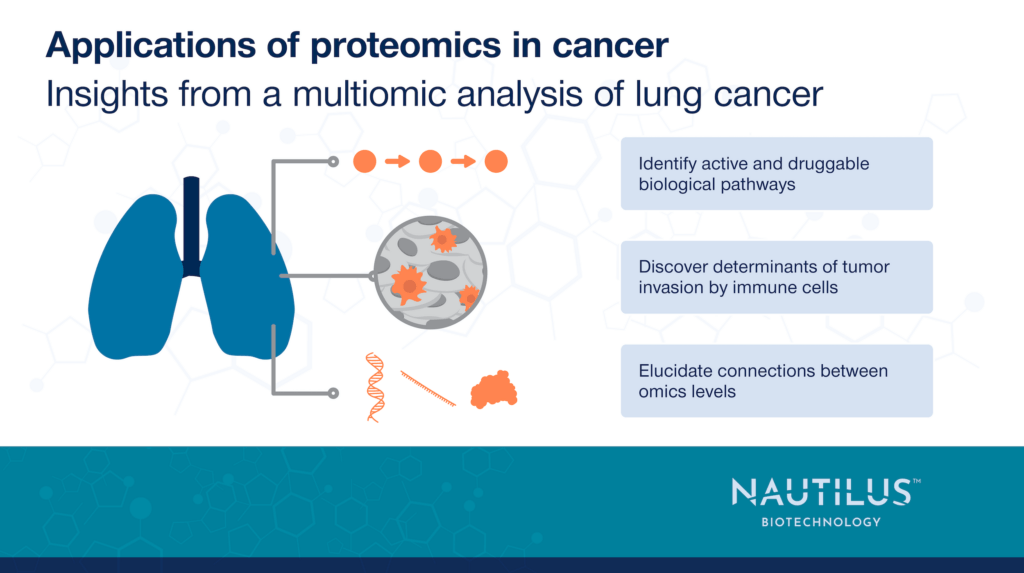
Applications of proteomics in cancer – Insights from a multiomic analysis of lung adenocarcinoma

Tyler Ford
November 7, 2023
The American Cancer Society estimates that over 100,000 people will die from lung cancer in the US in 2023. According to the CDC, 90% of these deaths will be linked to smoking, but many will not. As with all cancers, lung cancer is a mixture of different diseases and parsing the molecular causes behind lung cancer cases is critical to diagnosing and treating them effectively.
Toward this end, Gillette et al 2020. recently conducted a multiomic analysis of over 100 lung adenocarcinoma samples isolated from tumors and matching adjacent tissues. They combined genomics, transcriptomics, proteomics, phosphoproteomics, and epigenomics to identify mutated genes, altered methylation, differential RNA and protein expression, as well as proteoforms associated with lung cancer. With this extensive dataset, they were able to:
- Divide adenocarcinoma into multiomics-defined subtypes
- Infer the activity of mutant kinases on various substrates
- Discover cis and trans effects from copy number alterations
- Assess the effects of cancer-associated mutations on various biological pathways
- Associate particular mutations with immune evasion by tumor cells
- Identify potential drug targets and biomarkers
- Characterize pathways associated with adenocarcinoma in smokers compared to non-smokers
This work resulted in a treasure trove of data that can be leveraged for many future studies of lung cancer biology, biomarkers, and treatments. While this work made use of mass spectrometry-based proteomics, it provides an excellent example of the highly actionable insights proteomics, in general, can provide.
Below we cover a small fraction of these insights in more detail. We also highlight how we aim to make discoveries like these more accessible to people in many different research fields through the NautilusTM Proteome Analysis Platform. It is our goal to create a next-generation proteomics platform that makes it easier to generate data like that discussed here. We hope researchers can use our platform to advance that data to new realms of scientific discovery as well as the clinic.
View our animation to see how next-generation proteomics can fuel cancer research
Inferring the activity of kinases and identifying druggable pathways
The genomics work carried out by Gillette et al. 2020 identified fusions between kinases and other proteins in lung adenocarcinoma. Some fusions inactivated the kinases while others activated them, and some have been associated with lung cancer in the past. Examining just mRNA and protein expression would not capture how the fusions affect phosphorylation, so the researchers used phosphoproteomics to identify altered phosphorylation events indicative of kinase activity.
For example, fusions of the ALK kinase resulted in highly elevated phosphorylation of a variety of downstream targets. Not all such phosphorylation events necessarily drive cancer progression or will be druggable, but they point to promising avenues for further research that may lead to the identification of new drug targets.
Similar efforts identified copy number alterations as well as altered methylation patterns that impacted the expression of proteins both in cis and in trans. Some such mutations were in known cancer-associated proteins and, with their multiomic analysis, the researchers could trace the effects of these alterations to potentially druggable signaling pathways at the proteome and phosphoproteome levels.
Assessing immune activity in lung adenocarcinoma with multiomics
Tumors that have been invaded by immune cells are often associated with better cancer prognoses than those without invasion. The immune cells in these “immune hot” tumors have the potential to kill cancer cells and help prevent disease progression.
Gillette et al. 2020 used RNA-seq to group tumors into immune hot and immune cold subtypes. Then they used multiomics to find associations between immune invasion and particular mutations and potentially druggable signaling pathways. For instance, mutations in the STK11 protein were associated with low immune cell counts and high levels of proteins associated with an immune process called neutrophil degranulation. It is thus possible that manipulating this process in STK11 mutant tumors may increase the ability of immune cells to infiltrate tumors. As tumors with high levels of immune cell infiltration are not necessarily easily killed by those immune cells, similar analyses can help researchers identify pathways to manipulate for more effective immune cell activity even in these “immune hot” tumors.
Using multiomics to identify biomarkers indicative of underlying mutations in lung cancer
With their tumor samples and matched normal adjacent tissues, Gillette et al. 2020 could identify proteins upregulated in tumor cells with druggable mutations. They found high differential expression of proteins in TP53, EGFR, KRAS, and STK11 mutant tumors. Each of these mutant tumors had different sets of proteins that were differentially expressed and could be used as biomarkers. It was important to perform biomarker analysis at the protein level as opposed to the transcriptome level because mRNA expression did not correlate well with protein expression and may not predict the presence or activity of a druggable protein.
Advancing lung cancer research with accessible next-generation proteomics platforms
The ground-breaking work of mass spectrometry experts has made it clear that proteomics is critically important to truly understand biological processes. Nonetheless, we need more accessible and higher throughput next-generation proteomics platforms that can quickly assess the proteomes of many samples and sample types across many more labs. This will make it possible to efficiently advance the work discussed here and will enable researchers to revolutionize our understanding of and ability to treat not just lung adenocarcinoma, but all types of cancer. It is Nautilus’ goal to make such studies possible through the NautilusTM Proteome Analysis Platform. Learn more about our revolutionary platform here.
MORE ARTICLES