Applications of proteomics in cancer – Identifying potential precision medicines for cancer

Tyler Ford
July 18, 2024
The genomes of many cancers have been characterized but have limited utility to direct the development of precision medicines. Cancer comes with many mutations, and it can be difficult to determine which of them, if any, should be therapeutically targeted. Some mutations are not in coding regions, some have their greatest impacts downstream, some are in proteins that are difficult to target, and still others may not do anything.
For a more thorough look at the active, targetable proteins and pathways in cancer, researchers are leveraging multiomic analyses with a substantial focus on proteomics. These studies help researchers determine what biological pathways have abnormally high expression of their component proteins or active proteoforms. They directly show researchers what proteins can be targeted in cancer cells. They also make it easier to find multiple points of intervention by revealing full pathways that are active in cancer.
Liu et al. 2024 recently leveraged a multiomic analysis to reveal potential points of therapeutic intervention in small cell lung cancer (SCLC). The authors point out that, “Previous genomic studies revealed that inactivation of TP53 and RB1 occurs frequently in SCLC, but mutational profiling did not seem to establish molecular subtypes or identify actionable therapeutic targets.”
Below, we discuss some of the actionable findings reported by Liu et al. 2024 as a result of their multiomic analysis. We focus on biomarkers and therapeutic vulnerabilities important for the development of precision medicines. These findings provide an excellent example of the power of proteomics to enable targeted therapies.
Check out this animation to see how next-generation proteomics can fuel cancer research
Biomarkers of small cell lung cancer prognosis
Liu et al. 2024 identified two proteins with particularly strong associations with survival. High expression of HMGB3 was associated with shorter survival, while high expression of CASP10 was associated with longer survival. HMGB3 binds to DNA and plays roles in many processes including regulating transcription. It has been associated with worse outcomes in other cancers, but such associations have not been reported in small cell lung cancer. CASP10 plays a role in carrying out a form of cell death called apoptosis.
Liu et al. 2024 also used mRNA levels to divide their tumor samples into immune hot (high immune cell infiltration) and immune cold (less immune cell infiltration). The immune hot tumors were associated with longer survival than the cold. When the proteomes of the two immune subtypes were analyzed, the immune hot cells were positively correlated with immune related pathways, while immune cold cells were correlated with processes like DNA replication, cell cycle, and DNA damage repair.
Biomarkers like these, based on both individual proteins and on multiomics profiles, may make it easier for doctors to stratify patients into groups needing more or less aggressive treatment. Future studies that dive into the molecular details of the association between these biomarkers and survival may reveal additional ways to treat stratified patient groups with precision medicines. For example, patients with immune hot tumors may be more effectively treated with certain immunotherapies.
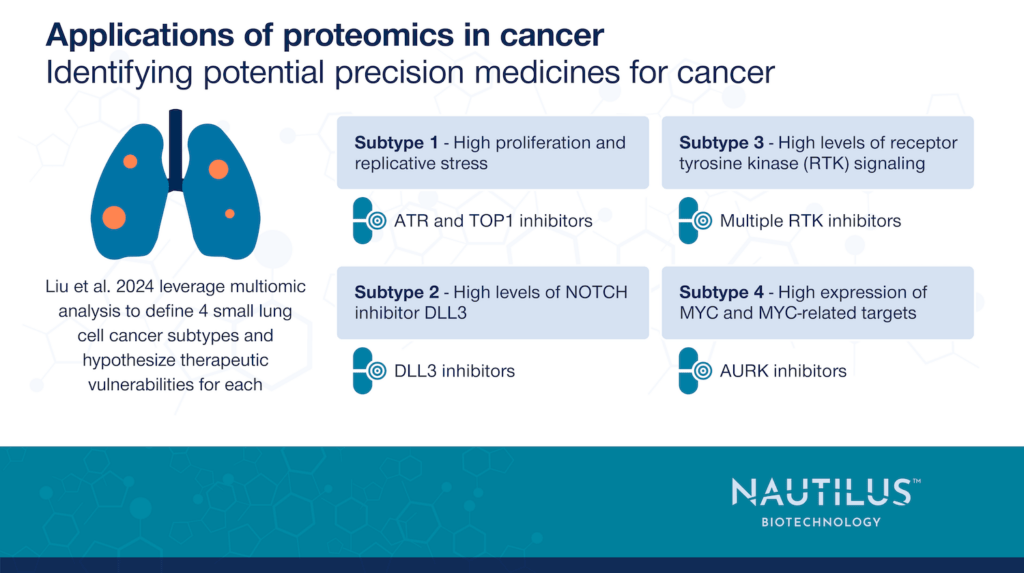
Small cell lung cancer subtypes with potential therapeutic vulnerabilities
Liu et al. 2024 separated the SCLC samples in to 4 multiomic subtypes with different potential therapeutic vulnerabilities:
Subtype 1 – Characterized by high proliferation and high replicative stress. The authors hypothesized they could target this subtype with ATR and TOP1 inhibitors. ATR is a kinase/DNA damage sensor that activates checkpoint signaling while TOP1 is a topoisomerase that relieves torsional tension in DNA during replication and transcription. Mouse xenograft models of subtype 1 responded well to ATR and TOP1 inhibition.
Subtype 2 – Characterized by inhibition of NOTCH signaling through high levels of the NOTCH inhibitor DLL3. NOTCH signaling normally plays a role in a wide variety of processes mediated through cell-to-cell communication. The authors believe therapeutically blocking DLL3 activity could effectively treat these tumors, but there were no DLL3-targeting agents available to them.
Subtype 3 – Characterized by high levels of receptor-tyrosine kinase signaling. Receptor tyrosine kinases are involved in many different functions including growth and development. The authors show that a multiple receptor tyrosine kinase inhibitor, anlotinib, prevents tumor growth in a mouse xenograft model of subtype 3 tumors.
Subtype 4 – Characterized by high levels of MYC expression and expression of MYC-related targets. MYC is a transcription factor that is frequently activated in cancer. The AURK kinases are involved in signaling downstream of MYC and stabilizing the MYC protein. Liu et al. 2024 predicated that AURK inhibitors could treat subtype 4 tumors. They used mouse xenograft models to show that AURK inhibition both slows subtype 4 tumor growth and inhibits the growth of subtype 4 tumors more than subtype 1 tumors.
A bright future for proteomics-enabled precision medicine
On their own, genomics studies can catalog mutations found in tumor samples of interest. Multiomics studies, on the other hand, enable researchers to establish links from genome to phenotype. As demonstrated by Liu et al. 2024, multiomics can thus provide critical insights into the protein pathways that directly mediate outcomes.
This work was particularly impressive because the researchers not only identified proteins and pathways that are potentially responsible for cancer outcomes, but also tested the effects of intervening with those proteins and pathways in animal models. It remains to be seen if the observations from these models will be transferrable to patients, but studies like these are sure to become more accessible as next-generation proteomics platforms with single-molecule and proteoform resolution become available. These will enable researchers to generate far more therapeutic hypotheses than they can with genomics alone. Studies like this one forecast a bright future for proteomics-enabled precision medicine.
MORE ARTICLES