Applications of proteomics in cancer – How proteomics complements genomic studies of colorectal cancer metastasis

Tyler Ford
July 23, 2024
Colorectal cancer (CRC) metastasis is a poorly understood process involving the migration of primary colorectal tumor cells to other parts of the body. It is a dire, yet common outcome of colorectal cancer diagnoses – 20% of diagnoses are metastatic and 25% of localized colorectal cancers will metastasize soon after diagnosis. When colorectal cancer metastasizes, it most often spreads to the liver.
A deeper comprehension of CRC metastasis from the colon to the liver could help scientists and clinicians better understand progression of the disease and also help inform prognosis and treatment. Toward this end, a recent study from the Roehrl Lab published in Cell Reports uses an assembly of omics tools to parse the molecular basis of this deadly process.
In their work, Tanaka et al. 2024 employ a multiomics approach leveraging genomics, transcriptomics, and proteomics to examine primary CRC samples and liver metastatic CRC samples along with normal colonic tissue and normal liver tissue. Their goal: to understand the driving forces behind primary tumor metastasis, which can help identify new drug targets to treat metastatic cancers.
Identifying distinct signatures of primary vs. metastatic CRC with the help of proteomics
A classic way to study cancer is to identify genetic mutations behind the development and progression of the disease. Such cancer-associated “driver mutations” include single-nucleotide variants, structural variants, and somatic copy number alterations. While identifying these mutations is useful, it doesn’t illustrate the complete story behind cancer progression.
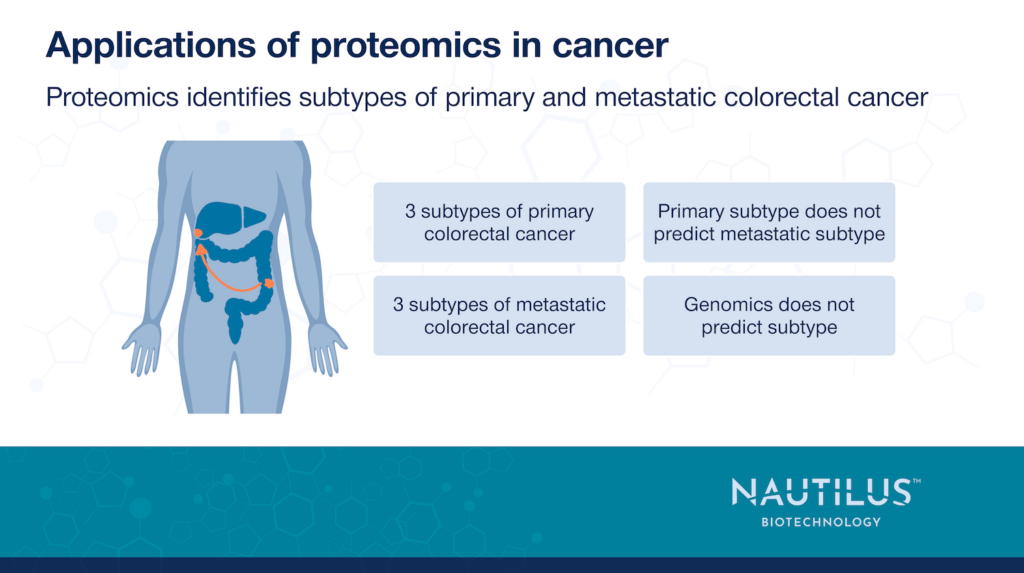
Tanaka et al. 2024 provide a great example of the limits of genomic data to explain CRC metastasis. When the authors examined the genomes of primary and metastatic CRC samples, they found some mutations were slightly enriched in metastatic CRC samples. However, they discovered that a purely genomic examination could not distinguish primary and metastatic CRCs.
That’s where proteomics comes in. With proteomic methods, the authors were able to reveal three distinct subtypes of primary CRC and three subtypes of metastatic CRC that have different hypoxia, stemness, and immune signatures. For example, proteomics found that some metastatic CRC samples had signatures of increased epithelial-mesenchymal transition and shifts towards anaerobic glycolysis. Furthermore, normal colonic tissue more closely resembled primary CRC than metastatic CRC. When the researchers examined the immune signatures of tumors, they found that metastatic CRC samples with high densities of T cells came from patients who had better outcomes.
Check out this video to see how next-generation proteomics can fuel cancer research
Gene mutations do not reflect proteome-based subtypes
After establishing the proteome-based subtypes, the researchers wanted to determine whether these subtypes were reflected by mutations in six genes commonly associated with cancer: APC, TP53, KRAS, PIK3CA, NRAS, and BRAF. While they did find associations between TP53 and two metastatic subtypes, KRAS and one metastatic subtype, and PIK3CA with a primary subtype, they could not detect any significant correlations in the mutation frequency of the other genes and other subtypes.
This finding demonstrates the value of proteomics. By looking at gene mutations alone, it would be difficult to distinguish one subtype from another. Primary and metastatic tumors exist in different environments so even though the mutations amongst them are not that different, proteomics reveals how the cells shift protein production based on their environment.
When the researchers examined matched primary CRC and metastatic CRC samples from the same patient, they also found that primary tumors from each of the subtypes could progress to any of the three metastatic CRC subtypes. In other words, the primary tumor subtype could not predict the metastatic tumor subtype. There were few genomic changes in major oncogenic genes upon progression from primary to metastatic CRC. This demonstrates yet again that metastasis is better reflected in the proteome than the genome.
Informing cancer diagnoses and treatments
This research demonstrates how important it is to look at the proteome, which can respond more rapidly to changes in real-time – for example, when cancer cells metastasize to the liver or respond to drug treatment. These proteomic changes have direct implications for the clinic and could help determine such things as:
- What tumors to analyze: This study shows that CRC tumors tend to have similar genomic profiles and their proteomes appear highly flexible as they can metastasize to any of the three subtypes. Therefore, it’s important to evaluate metastasized tumors in addition to primary tumors as the primary tumors may not accurately reflect the proteome of metastasized tumors in the same patient.
- What treatments to pursue: Proteomic subtypes could better determine how to treat CRC than predictions based on genomics. For example, stemness-high CRC tumors had highly upregulated drug transporters that could reduce the effectiveness of current drugs. Other, hypoxia-high metastatic CRC tumors had increased vascularization and epithelial-to-mesenchymal transition so targeting these processes could be effective treatments in this subtype. It’s also possible for one tumor but not another to respond to a particular drug in the same patient, further underscoring the need to examine the proteomes of multiple tumors.
Next-generation proteomics for studying cancer metastasis
Tanaka et al. 2024 show just how useful it is to study cancer metastasis using a multiomics approach. While there are numerous gene mutations associated with cancer, proteomic studies help scientists understand the biology of tumors at a particular time and place. These studies thereby provide information that it is critical for effective cancer treatment in realtime.
Next-generation proteomics platforms like the Nautilus™ Proteome Analysis Platform can help researchers studying cancer metastasis by providing wide dynamic range, single-molecule resolution, and robust and reproducible analyses.
Learn more about the Nautilus Platform here and check out this episode of the Translating Proteomics podcast to learn how we can leverage multiomics to study “Biology in Space and Time”:
MORE ARTICLES