
Applications of proteomics in agriculture – Disease resistance

Tyler Ford
July 20, 2023
Disease isn’t just a problem for humans. Fungi, viruses, and bacteria infect plants too, and the diseases they cause can wipe out crops, causing financial ruin for farmers and even imperiling food supplies.
In the third part of our Applications of proteomics in agriculture series, we’ll show how proteomics is deepening our understanding of plant pathogen defense. Plants and microbes are engaged in an eons-long showdown involving sophisticated strategies of chemical warfare on either side. Many pathogens have adapted to infect specific species of plants, and the plants have responded with an arsenal of defensive chemicals and strategies meant to fend them off.
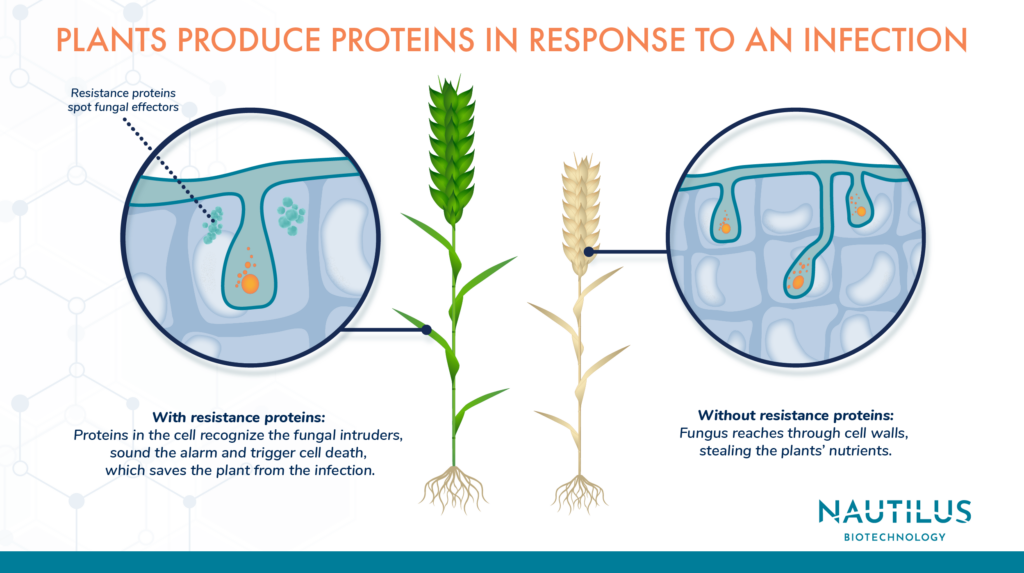
Proteins play a role on both sides of this conflict. Pathogens use them to slip inside plant cells and take over. Plants use them to counter invasions, befuddle attackers, and even shut down pathogens entirely.
Researchers can use proteomics to identify proteins involved in plant-pathogen warfare and better understand the molecular mechanisms that underlie resistance to specific pathogens. This information could lead to the development of new strategies to control plant diseases, including the creation of new disease-resistant crops.
Fortifying plant defenses with proteomics
Proteomic analysis has been used to identify the plant proteins that are involved in resistance to fungal, bacterial, and viral pathogens in some of the most important crops. Wheat is the most widely-grown crop in the world by land area, but the fungal pathogen Fusarium graminearum, which causes Fusarium head blight, can cause up to 70% reductions in yield. The fungus causes the heads of wheat plants to bleach and die, and can produce a toxin called deoxynivalenol that can harm both humans and animals.
To try and solve this problem, researchers have used proteomics to identify the proteins that help wheat defend against Fusarium head blight. Studying 7,910 proteins across three wheat varieties with different levels of resistance, they found two specific proteins were missing in the most susceptible variety. These two proteins (one related to mitochondrial reactive oxygen species and the other known to be related to fungal resistance) could be important for defense against Fusarium head blight. Nonetheless, 98% of identified proteins were present in all samples, indicating that, for most proteins, their relative abundance, as opposed to simple presence or absence, may drive wheat resistance to this fungus. Researchers may be able to develop effective strategies to stop fungal disease in wheat and other plants with a more comprehensive understanding of their proteomes.
On the flip side, proteomics isn’t just applicable to the plants affected by these diseases. It can also be used to better understand the tools that pathogens use to infect their hosts. In a study published in Genetics and Resistance, researchers used proteomics to look at small cysteine-rich proteins secreted by the F. graminearum fungus during infection. They found proteins that are highly expressed by the fungus when it infects plants and may cause disease. Stopping the fungus from making these proteins, or giving plants the ability to counter them, could stop infections in their tracks.
Proteomics probes plant viruses
Scientists have made strides in using proteomics to counter viral infections in plants as well. Viruses have striking differences in their biology compared to bacteria or fungi and, critically, cannot replicate on their own. Instead they rely on host protein machinery to synthesize their genetic material and produce viral proteins. Proteomics is beginning to illuminate the processes that help plants resist infection from these peculiar pathogens.
For example, Pea seed-borne mosaic virus infects the common pea plant Pisium sativum. In a 2017 paper in the Journal of Proteomics, scientists compared the proteomes of resistant and susceptible pea plants and identified 116 different plant proteins that responded to viral infection. Some of these proteins were involved in known resistance pathways like carbohydrate metabolism and amino acid biosynthesis. But they found novel proteins too, including a lipid metabolism protein affecting how proteins are moved around the cell and involved in wound responses. The ability to assess the whole proteome at different stages of infection helped them find numerous proteins involved in both resistance and susceptibility, which could aid the design of disease resistant plants in the future.
New tools for more impactful proteomics applications in agriculture
Just as in medicine, proteomics has become an increasingly important tool in agricultural research, providing insights into the molecular mechanisms underlying plant and animal growth and health. In plants, proteomics can be used to study the molecular basis of stress adaptation and tolerance, leading to the development of crop varieties that can grow in poor soil, high temperature, and low water. Proteomics can also help researchers decipher the complex chemical warfare waged between plants and pathogens, revealing the signals, processes, and responses used by each side.
In all cases, knowledge from proteomics research will empower scientists to continually improve crops to meet the challenge of feeding a growing world population in an ever-more-challenging climate. New advances in proteomics technologies, such as the Nautilus Proteome Analysis Platform, are designed to improve the sensitivity, dynamic range, accessibility, and efficiency of proteomics studies so that more researchers can deeply probe important biological process in agriculture and beyond.
Listen the Translating Proteomics podcast for a fascinating discussion of the applications of proteomics
MORE ARTICLES
