
Advantages of the Nautilus Proteome Analysis Platform – Wide dynamic range

Tyler Ford
January 16, 2024
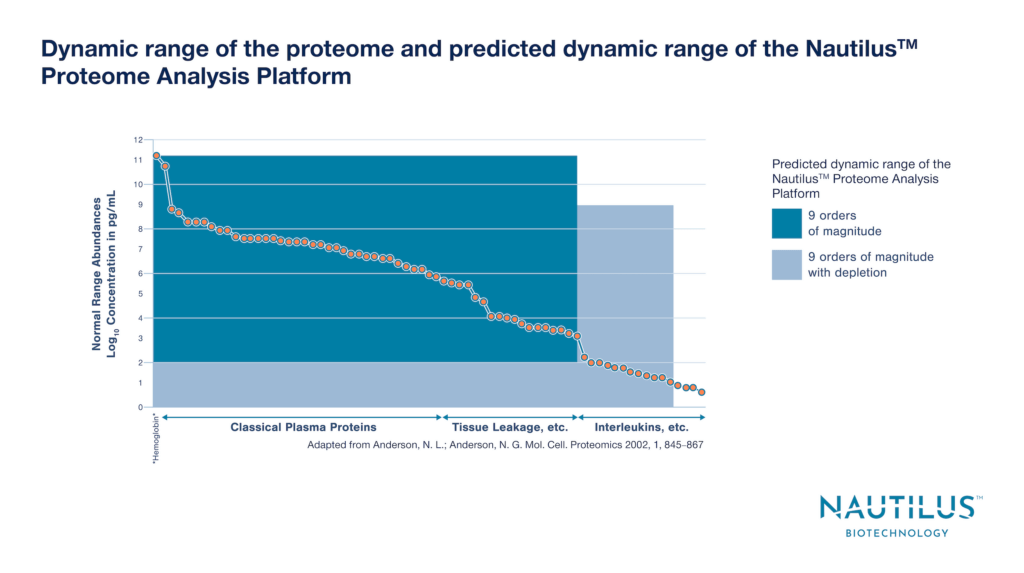
Protein abundances lie across a wide dynamic range in the proteome. For instance, proteins in the human plasma proteome have abundance differences spanning roughly 12 orders of magnitude. Both high and low abundance proteins serve important functions, and it is essential to measure them all in a comprehensive proteomic analysis designed to provide mechanistic insights into cellular functions and organismal behaviors.
Learn more about plasma proteomics.
The NautilusTM Proteome Analysis Platform is designed to analyze up to 10 billion single protein molecules in a single run. This is expected to provide unprecedented dynamic range that will cover substantively the entire proteome.
There are two major components of our process and platform that are expected to enable such a wide dynamic range:
- Massive arrays with 10 billion landing pads for single protein molecules.
- Protein library preparation that ensures super-Poisson loading of our array and maximizes the number of molecules analyzed per run.
Creating massive arrays to match the scale of the proteome
The first component is simple in concept but was only recently possible. Using nano-etching techniques, we can reliably create arrays that have 10 billion landing pads for single protein molecules. In addition, we can coat these intricately patterned arrays and give each landing pad a charge that attracts scaffolded proteins as described in the next section. These arrays are the culmination of years of nanofabrication research and engineering. We are incredibly lucky to live in a time where their creation is not only possible but practically feasible at scale.
Making single protein molecules uniform using DNA scaffolds to enable super-Poisson loading
In the past, researchers often used limiting dilution to isolate single molecules on arrays. However, limiting dilution results in Poisson loading. This means some array landing pads have single molecules, but only a small number of pads are occupied, and 50% of the occupied pads have more than one protein.
If you could be sure that only one protein would fit on every landing pad on an array, you could achieve single protein occupancy across the whole array without limiting dilution. However, even with the most modern fabrication techniques, proteins are too small and too diverse in size to reliably achieve single-molecule loading of every landing pad.
Our solution is to scaffold proteins to DNA nanostructures and thus give them a relatively uniform size. We use click chemistry to attach single, intact, and denatured proteins to the DNA nanostructures and generate single-molecule protein libraries. The nanostructures are designed to ensure only one can fit on each landing pad. The negative charge of the DNA nanostructures also increases their affinity for the positively charged landing pads and helps ensure that the scaffolded proteins load with the correct orientation. As a result, we achieve super-Poisson loading where the majority of our 10 billion landing pads are occupied by single protein molecules.
Watch this video for a visual dive into the dynamic range of the Nautilus Platform
Quantifying proteins across the dynamic range of the proteome
With our 10 billion landing pad arrays and proprietary protein library preparation techniques, we can achieve super-Poisson loading that makes it possible to probe up to 10 billion proteins at the single-molecule level. Thus, our platform is designed with an unprecedented up to 9 orders of magnitude dynamic range. With this dynamic range, we expect to be able to identify proteins across substantively the entire proteome. We believe this will enable incredible advances in our mechanistic understanding of cell and organismal function that will lead to a revolution in basic research, clinical development, and beyond.
To learn how our single-molecule protein identification process works, check out our PrISM preprint and tech note.
For more information on our arrays, check out our array pre-print.
MORE ARTICLES
