Biology researchers have learned much about the intricate inner workings of cells using classic molecular biology, biochemistry, and genetics techniques. Generally, these techniques minimize biological complexity by looking at only a small sub-component of a cell in a highly controlled experimental setting. For instance, researchers might break a small part of the cell (say a single protein) and see what effect that has on the behavior of the cell or organism. Although they have brought us far, such techniques are by their nature reductionist and make it difficult to assess the many interactions occuring in living things at any moment in a holistic way.
To truly understand how living things function in a mechanistic way that can be leveraged for applied purposes, researchers need to bridge the gap between small cellular components and broad organismal behaviors. They need to map out the intricate networks of interactions that connect cellular parts to organismal functions. Proteomics can enable them to do so.
Proteins carry out the vast majority of cellular processes and a cell’s proteome, the dynamic composition of its full set of proteins and their associated distribution throughout the cell, determines cellular identity, form, and function. Proteomics technologies aim to reveal the abundances and locations of all proteins in cell samples right now. The dynamic patterns of these ever-changing proteomes comprise thousands to millions of fluctuations in protein levels, protein modifications, and protein distribution throughout the cell, and can help reveal the networks of protein interactions that are perturbed when cells experience various stimuli.
In this series, we discuss 3 types of research that can be accelerated by advanced proteomics technologies. This is far from an inclusive list and focuses on non-clinical research with a nod to clinical applications that can result from these research areas. As we get accessible, high-throughput proteomics technologies into the hands of researchers around the world, they’ll develop myriad applications that will reveal much more about the inner-workings of living things than we ever thought possible.
Learn how Professor Kathryn Lilley and others are leveraging proteomics to learn more about protein function:
Mapping the proteomic landscape of the cell surface
Cells are not homogenous bags of proteins. Instead, proteins are distributed to discrete locations throughout the cell where they act in coordination with other cellular components to carry out specialized processes. Proteins found at the cell surface are often inserted into the “plasma membrane,” the fatty barrier separating the inside of the cell from its surroundings. These “membrane proteins” are particularly important because they make it possible to interact with and relay information about a cell’s surroundings. While membrane proteins are traditionally difficult to study, new proteomics technologies will make it easier for scientists to learn more about these incredible molecular machines.
Membrane proteins have varied and fascinating functions. For example, some proteins on the surface of immune cells detect parts of infectious bacteria. Once detected, these proteins trigger a cascade of events that can result in the consumption and later destruction of the bacteria. In another interesting example, proteins on the surface of nerve cells control the flow of ions in and out of the membrane. They enable our nervous systems to transmit highly controlled electrical signals. Still other proteins embedded in the surface of retinal cells detect specific wavelengths of light, a function that ultimately gives us color vision.
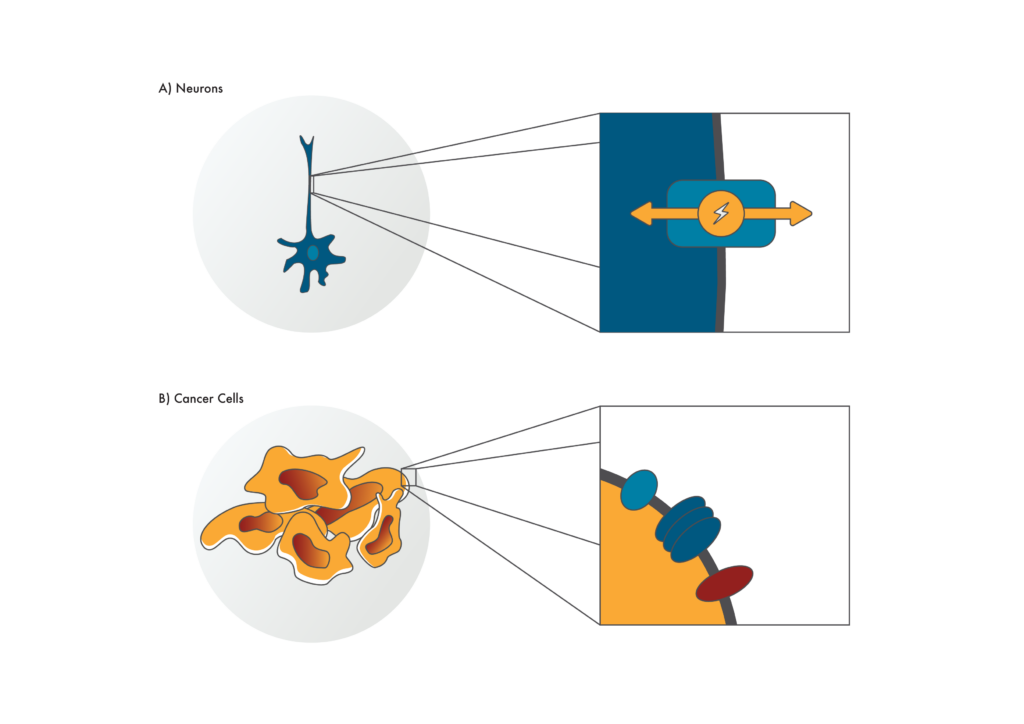
Figure 1. Cell surface proteins
Cell surface proteins or “membrane proteins” have a variety of fascinating functions such as (A) enabling neurons to send electrical signals. While they can be hard to isolate and study, if researchers can (B) find membrane proteins specific to cancer cells, these may make good drug targets.
Unsurprisingly, membrane proteins are a fascinating subject for scientists wishing to understand how our bodies work, but they are also incredibly important in the clinic. Membrane proteins make useful drug targets because they’re easier to reach than proteins buried inside the cell. They’re also accessible to large, programmable biological therapeutics like antibodies and engineered cells.
Unfortunately, membrane proteins can be very difficult to study. They are not usually soluble in water and can be hard to purify from the fatty barriers where they reside. In addition, they sometimes stick to the materials scientists use to store and transport them. As a result, scientists often have to use strong chemicals to break apart the membranes housing these proteins and keep them from sticking to their containers. These chemicals are generally incompatible with standard protein analysis techniques.
These characteristics make membrane proteins difficult to study not only on an individual level but also at the level of the proteome. With traditional proteomics techniques, scientists usually use enzymes to cut proteins into smaller, more manageable chunks. Membrane proteins contain few cut sites and the strong chemicals used to pull them out of the membrane disrupt the enzymes’ cutting activity. Scientists have come up with a variety of clever ways to get around these issues, but they are usually complicated and time-consuming.
An additional problem for standard proteomics techniques is membrane protein abundance. Membrane proteins are generally low abundance and the signals they generate on standard proteomics platforms can be drown out by other proteins. This is because standard proteomics platforms don’t look at individual proteins in isolation. Instead, they identify small protein chunks from many different proteins. Various kinds of analysis software then piece together the identities of the full proteins from the small chunks. This is much like putting together a very complicated puzzle. High abundance proteins generate many small chunks that can obscure the identities of the low abundance proteins. Scientists can isolate membrane proteins and analyze them separately, but this can bias which proteins they ultimately see and complicate the experimental process.
Nautilus’ technology is designed to get around these issues in a number of ways. First off, scientists don’t need to break apart their proteins before analyzing them on the Nautilus’ platform. The platform is also designed to be compatible with the strong chemicals used to pull proteins out of membranes. Special isolation techniques are not required. Finally, full-length proteins are analyzed individually on the platform. This is expected to give the platform higher sensitivity and make it much less likely that signals from high abundance proteins will drown out membrane proteins.
Thus, we hope the Nautilus platform will open the proteomic landscape of the cell surface to scientists. We aim to make it so they can use just about any chemicals they wish before analyzing membrane proteins and they’ll also get a full view of most of these proteins, regardless of their abundance.
With this landscape in view, scientists are expected to be able to develop a clear understanding of how cells interact with their surroundings. For instance, they’ll be able to determine what proteins stud different types of cells and, by associating particular membrane proteins with known cell-cell interactions, generate new hypotheses about how these interactions function at the molecular level. They’ll also be able to probe how surface proteins change in various states of health and disease. For example, by studying the proteins found on the surface of cancer cells compared to healthy cells, they may find cancer-specific proteins. Thus, they’ll be able to target these proteins with cancer-specific drugs – something that is only possible for a few types of cancer now.
Overall, advances in proteomics technology are expected to enable researchers to answer a seemingly basic question: what proteins are found on the cell surface? The answers will advance our understanding of how the body works and improve health for many.
Want to read the full series now? Download the “Accelerating basic research with proteomics” white paper here.
MORE ARTICLES



