
A brief introduction to PrISM – The theoretical framework underpinning the Nautilus platform

Chris Blessington
September 9, 2022
With Nautilus’ founding, we set out to develop a technology that could deliver a simple but information-rich readout of the abundance of tens of thousands of proteins from just about any sample of interest with minimal sample prep and hands-on time. The theoretical framework underpinning our platform is called Protein Identification by Short epitope Mapping (PrISM).
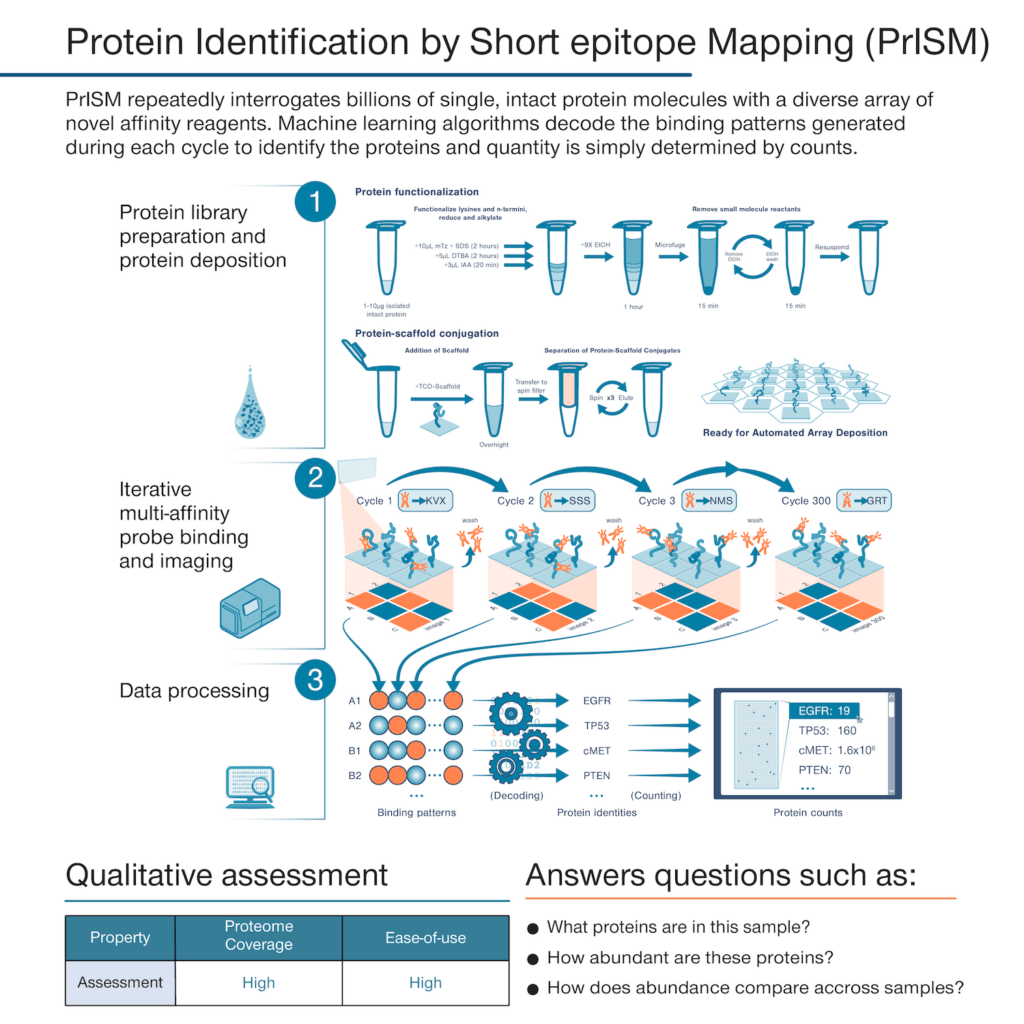
With this technique, rather than use highly specific affinity reagents that bind to individual proteins, we make use of many affinity reagents that bind to short sequences of amino acids with high affinity and low specificity. These reagents are designed to stochastically bind many different proteins. If one were to add just a few of them to a sample and monitor binding, they would not identify any individual proteins with confidence. Instead, PrISM is designed to monitor how hundreds of these affinity reagents bind to individual proteins at the single molecule level. This generates binding patterns for each protein molecule and then the patterns can be decoded by machine learning algorithms configured to determine the identity of each protein molecule.
See our recent bioRxiv pre-prints for more information on our affinity reagents, the framework for Protein Identification by Short Epitope Mapping (PrISM), and the development of our single-molecule protein arrays.
MORE ARTICLES
