
Proteomics and the development of precision medicines against cancer (Part 2)

Tyler Ford
February 21, 2023
Cancer is a heterogenous mixture of diseases characterized by, among other things, the abnormal, uncontrolled growth of cells derived from otherwise healthy tissues (Hanahan 2022). Although cancer cells sometimes grow into balls of cells and stop there (so-called benign tumors), often they gain the ability to disperse throughout the body, seed the growth of other tumors, disrupt the function of a variety of organs, and ultimately kill patients. In this series of blog posts, we discuss how proteomics can drive the creation and use of “precision medicines” that target the processes enabling cancer growth and stop cancer in its tracks.
Want to read the full series now?
Proteomics and cancer diagnostics
When a patient presents with an abnormal, possibly cancerous growth, doctors may begin their investigation into whether or not it’s dangerous by taking a biopsy, a sample of the growth. Often, they’ll analyze the sample under a microscope and look for physical features associated with uncontrolled growth. Increasingly, they’ll also sequence the DNA from the biopsy and thereby identify any mutations known to be associated with certain types of cancer.
This information may be enough to tell physicians whether or not the growth is dangerous, but only rarely do these efforts provide certainty about the precise molecular cause of the growth. In the rare cases where the molecular cause is obvious from DNA sequencing, doctors may be able to prescribe their patients with treatments that directly target the proteins involved. This can be highly beneficial for patients and, in fact, studies have shown that such “precision medicines” are more effective than treatments chosen without the aid of sequencing (Morash et al 2018).
Currently this is only possible for a small subset of cancer patients because mutations in DNA do not directly tell scientists how the encoded proteins are affected. A mutation is generally necessary for an effect but not all mutations are driver mutations. Using advanced proteomics technologies, physicians can get information that directly shows how proteins are perturbed in cancer cells. While traditional proteomics technologies don’t always have the sensitivity, accessibility, or throughput required to analyze proteins from small patient samples and large numbers of patients, up-and-coming technologies are designed to have much higher sensitivity and aim to catalog the vast majority (95% in Nautilus’ case) of the human proteome in as little as 24 hours.
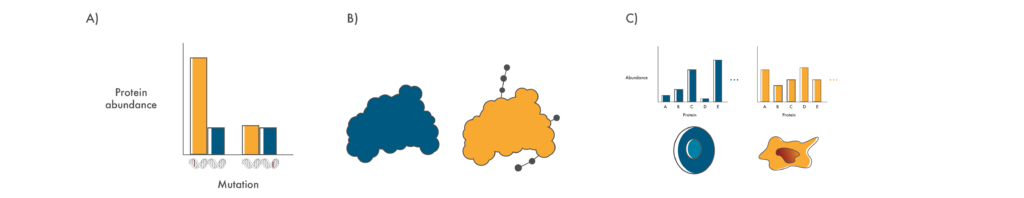
Questions that proteomics can help answer
A) Is the protein encoded in a mutated gene produced in cancer cells in a significant amount? B) Does a mutation result in changes to post translational modifications on proteins? C) Does a mutation result in wide-scale proteome changes in cancer cells?
This will give researchers a variety of actionable pieces of information including:
Whether proteins encoded in mutated genes are produced in any significant amount.
Cancer cells often contain mutations in multiple genes and it is far from obvious which of these might be mechanistically involved in the cancer. Using proteomics, researchers can check if any of these proteins are actually produced and assess their production relative to matched healthy cells. Those that are expressed at higher levels may be good drug targets while those expressed at lower levels might give insights into the molecular pathways involved in that particular cancer.
Knowledge of proteome-wide post-translational modifications.
Although there are roughly 20,000 protein-coding genes in the human genome, the proteins encoded in these genes can be modified in many ways once they are “translated” or produced from this information. These post-translational modifications (PTMs) have many functional impacts on proteins. They can make proteins more active, change where they’re located in the cell, and even mark them for destruction. With their many roles, PTMs have far-reaching impacts on cancer.
In particular, a PTM known as “phosphorylation” is often involved in activating signaling pathways that cause cells to grow. By using proteomics to discover what proteins are phosphorylated, researchers can associate particular signaling pathways with cancer initiation and progression (Mani et al 2022). They can also inhibit the phosphorylation events involved to halt cancer progression.
The production and activity of many proteins can also be altered by the post-translational modification known as acetylation. This PTM involves the addition of a small chemical group to parts of proteins. For DNA-scaffolding proteins called histones, this can result in the DNA being more or less accessible to other proteins that read and decode it. When acetylation causes a stretch of DNA to be more accessible, the proteins in that stretch can be produced in higher amounts and vice versa. Acetylation of other proteins can also alter their activity and lead to changes in cellular function. In fact, changes in acetylation are associated with characteristics that can drive cancer, that can stratify cancer severity, and that can direct physicians to specific treatments (Harachi et al 2021, Zhang et al 2016). If researchers can use proteomics technologies to associate more acetylation events with specific cancer cell functionalities, they may also be able to design compounds that inhibit these acetylation events or modify the activities of acetylated proteins.
A genome-agnostic assessment of changes in protein abundance.
In the above examples, proteomics technologies are framed as a means to complement genomics. However, proteomics has a large role to play on its own. By comparing the proteomes of cancer cells to the healthy cells they’re derived from, researchers can identify proteins with altered abundance regardless of the underlying genomic mutations. In addition, by comparing the proteomes of cancer cells from patients with varying degrees of aggressive disease, they can independently associate particular “proteomic profiles” with levels of severity. Using this information, physicians can treat their patients more or less aggressively. For instance, they might be more willing to use a treatment with particularly strong side effects if a patient has a very severe cancer and poor prognosis.
Looking at proteomic data agnostic to genomics data can also be useful because a variety of non-genomic factors can alter cancer cell behavior. For instance, cancer cells can often resist attacks from immune cells and even produce compounds that tamp-down the immune response. Production of the proteins involved in these interactions can be induced by genetic mutations, communication with other cells, and conditions in the tumor’s environment. Even when immune evasion is genetically based, the changes in protein expression that enable it won’t necessarily be obvious from the initial mutations alone. Thus, by looking at changes in protein expression indicative of immune evasion instead of looking at the genome, researchers can devise ways to effectively give the immune system a boost and kill cancer cells (Hanahan and Weinberg 2011, Spranger and Gajewski 2018).
Similarly, cancer cell interactions with the microorganisms that live on and in us, the microbiome, are increasingly being recognized as important factors in cancer progression (Hanahan 2022). Although there are not always genomic mutations associated with some of these interactions, researchers can use proteomics to look for protein-based signatures of them.
Overall, proteome-based cancer diagnostics can provide researchers and doctors with far more clinically-actionable information than diagnostics of the past. In the next post in this series, we’ll discuss how proteomics studies can help researchers and doctors develop new cancer treatments.
Want to read the full series now? Download the “Proteomics and the development or precision medicines against cancer” white paper here.
View our animation to see how next-generation proteomics can fuel cancer research
MORE ARTICLES
