
Spatiotemporal changes to the proteome in Angelman syndrome – Applications of proteomics in neuroscience

Tyler Ford
August 22, 2024
Angelman syndrome is a neurological disorder that leads to developmental delays, intellectual disability, and a range of other symptoms. It is a rare genetic condition caused by mutations in the UBE3A gene with a prevalence of 1/20,000 – 1/12,000 live births. Because the paternal allele of UBE3A is normally silenced (imprinted), mutations in the maternal allele are sufficient to cause the disorder (See Madaan and Mendez 2024 for more information).
Many people with Angelman syndrome have a normal paternal allele for UBE3A, and drugs designed to activate this allele are currently in clinical trials. Indeed, studies in mice have shown that reinstating UBE3A protein levels soon after birth can largely rescue the physiological effects of the disease. The impact is much less pronounced if UBE3A is reinstated in adulthood (Silva-santos et al., 2015).
To better understand the biology underlying Angelman syndrome and the intricacies of rescue, Pandya et al., 2022 leveraged proteomic and targeted protein analysis across mouse, rat, and human cell line-based models of the disease. Their work not only identified biological processes prominently impacted by the disease and its rescue but also highlighted varying degrees of rescue from UBE3A reinstatement at different developmental time points. Finally, their work showed a canonically metabolic enzyme called transketolase may play a surprising role in the disorder.
Watch this animation to learn how next-generation proteomics platforms can fuel neuroscience
Proteomic analyses of mouse, rat, and human Angelman syndrome models reveal aberrations in proteostasis and synaptic function
Pandya et al., 2022 used mass spectrometry to quantify proteins with altered abundance in UBE3A knockout mice and rats. In both cases, the rodents displayed developmental delays and other symptoms of Angelman syndrome. At the level of the proteome, the rodents had significant changes in proteins involved in:
- tRNA synthetase pathways – The levels of tRNA synthetase pathway proteins had both increases and decreases compared to controls. tRNA synthetases are essential components in protein production and alterations in these pathways could have wide-ranging effects.
- Proteasomal pathways – The levels of these proteins were generally increased compared to controls. Proteasomal proteins coordinate protein degradation and these too could have broad-ranging effects on biological function.
- Synaptic factors – Synaptic proteins were both increased and decreased in the rodent models compared to controls. Synapses form the hub of signal transmission between neurons and alterations in these proteins could have broad impacts on neurological function.
Similar patterns of protein expression were seen in iPSCs derived from Angelman syndrome patient neurons compared to controls.
Spatiotemporal proteomics in rodent models of Angelman syndrome
In the rats, the researchers also looked at differences in patterns of protein abundance across brain regions. While the patterns above were generally recapitulated across regions, they did observe broad proteomic differences between regions indicating differences in their development. Further work could elucidate how these differences impact development.
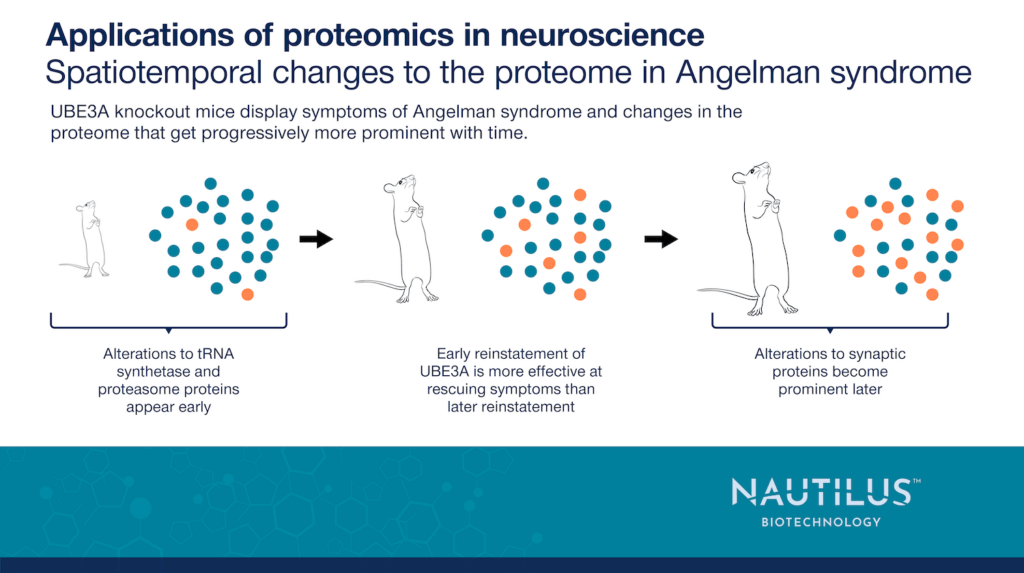
Pandya et al., 2022 additionally looked at changes in mouse protein expression at birth, during adolescence, and in adulthood. Changes in tRNA synthetase and proteasome pathways are prominent at birth, but changes in synaptic proteins became gradually more prominent over time. This may reflect a gradual impairment of brain development stemming from more immediate impacts on tRNA synthetase and proteasomal pathways.
Discrepancies between Angelman syndrome proteomic and physiological rescue at different developmental time points
Pandya et al., 2022 also set out to determine how changes in the proteome correlate with the rescue of physiological function brought about by reinstating UBE3A expression at different developmental time points. To do so, they used a tamoxifen-inducible UBE3A mouse line and reinstated UBE3A expression at adolescence and adulthood. Early reinstatement rescues physiological function while late reinstatement does not. Interestingly, however, reinstating UBE3A during adolescence rescues many impacts on the proteome, while reinstating during adulthood partially rescues proteomic changes. This may indicate that further intervention is necessary with later UBE3A reinstatement.
Proteomics identifies a surprising target of UBE3A
Across all their models, Pandya et al., 2022 observed that the abundance of an enzyme called transketolase increases in Angelman syndrome. Further analysis showed that nuclear levels of this protein particularly increase in neurons. Although transketolase nuclear localization had been reported in non-neuronal cells prior to this work, given that transketolase canonically catalyzes a reaction in the pentose phosphate pathway, and this pathway occurs in the cytoplasm, this prominent nuclear localization points to a non-canonical role for transketolase. Pandya et al., 2022 further showed that UBE3A likely directly ubiquitinates transketolase in the nucleus.
The findings that transketolase is consistently up-regulated across Angelman syndrome models, and that it is likely a direct target of UBE3A indicate that transketolase plays an important role in Angelman syndrome. More work is necessary to determine this role, but these results are a great demonstration of the insights that can come from proteomics.
Implications for future work on Angelman syndrome
The results presented by Pandya et al., 2022 have a variety of implications for future academic and clinical research into Angelman syndrome. On the academic side, it will be interesting to see how modifying levels of proteins downstream of UBE3A impact neuronal networks across the brain. In addition, studying the function of transketolase in neuronal nuclei may reveal interesting new functions for this enzyme. In the clinic, these results reinforce previous studies suggesting early reinstatement of UBE3A expression has greater impacts than later reinstatement. They also point to some downstream proteins that may need further intervention to rescue the pathology of older Angelman syndrome patients.
Overall, this work represents a thorough integration of proteomics, in vivo models, and in vitro models. This kind of work requires researchers with various types of expertise and experimental skills to collaborate extensively. We hope next-generation proteomics technologies like the NautilusTM Proteome Analysis Platform will make integrative and collaborative efforts like this one far more accessible.
Find more applications of proteomics in our Proteomics and neuroscience eBook.
MORE ARTICLES

