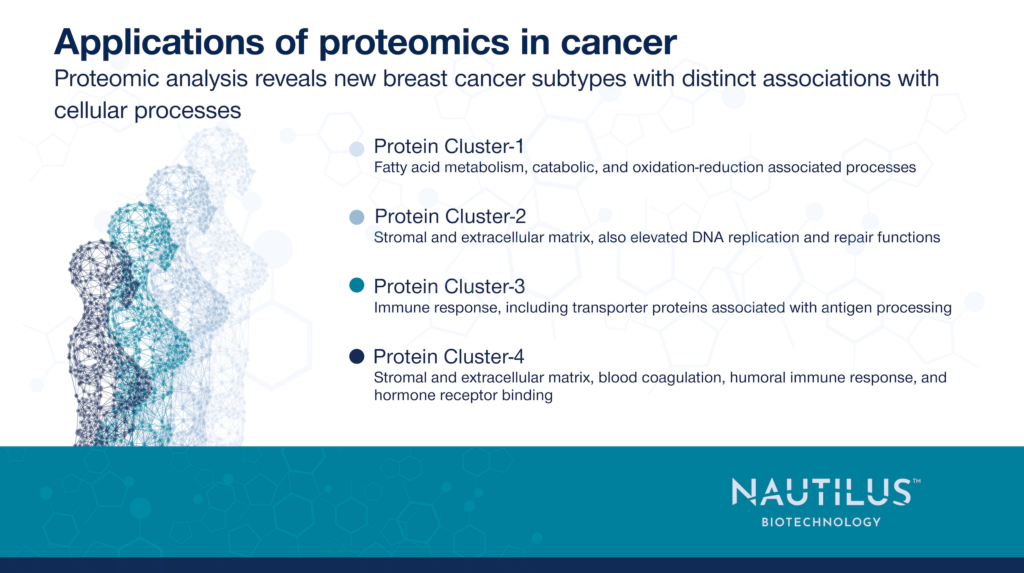
Applications of proteomics in cancer – Proteomic analyses can reveal new breast cancer subtypes

Tyler Ford
October 19, 2023
In the U.S. alone, more than 260,000 people are diagnosed with, and more than 40,000 people die of breast cancer every year according to the Centers for Disease Control and Prevention. Finding new ways to assess and treat the disease could potentially save thousands of lives every year. Toward this end, a recent proteomic analysis of breast cancer samples has revealed new subtypes of this common cancer. This work provides researchers with a more precise understanding of the disease that can hopefully lead to more effective diagnostics and treatments.
Published in Nature Communications in 2022, the work was conducted by a team of researchers from the Morin Lab at the University of British Columbia. They used proteomics to analyze 300 breast cancer specimens. Their analysis revealed the specimens could be grouped into distinct cancer subtypes based on their proteomes. Using information from a biobank, the researchers were able to see that patients with certain proteome-defined cancer subtypes had similar treatment outcomes.
Researchers can use these different breast cancer subtypes to help reveal what makes certain cancers deadly and why some patients respond to treatments. Proteomic analyses like this one demonstrate the mountain of actionable information in the proteome and showcase the value of bringing that information to light.
Finding more predictive breast cancer subtypes
Breast cancer arises when cells in the breast proliferate uncontrollably. Each specific instance of breast cancer can look different, depending on things like a person’s genetics, and what mutations led to the cancer. For example, one way researchers classify breast cancers is a test known as the Prediction Analysis of Microarray 50 (PAM50) that looks at RNA expression from 50 genes known to be associated with the disease.
Knowing which breast cancer subtype a patient has helps show doctors which treatments may work best and helps them deliver a prognosis. Subtypes based on RNA aren’t perfect, though. Gene expression at the RNA level does not always correlate well with protein expression, and there are several processes, like mRNA splicing, as well as post-translational modifications that can cause proteins to look different from the genes that encode them. Ultimately, looking directly at proteins instead of transcripts may be a better way to differentiate one cancer from another.
View our animation to see how next-generation proteomics can fuel cancer research
Using proteomic analysis to understand breast cancer
For a better look at the diversity of breast cancer, the researchers turned to discovery proteomics. This technique uses proteomics tools like mass spectrometry to analyze all of the proteins in a given sample, showing researchers protein identities, protein quantities, and what protein variations, called proteoforms, exist in the sample. Proteomic analyses like this are fundamental tools of modern biology.
Using mass spectrometry, they analyzed the proteins in each breast cancer specimen, ending up with a list of 4,214 proteins that they were able to quantify in every sample.
The researchers’ proteomic analysis enabled them to subdivide the samples into four unique groups that overlapped with canonical subtypes, but didn’t mirror them. The researchers also used protein-level data to identify biological pathways associated with different prognoses. For example, they found that proteins indicative of a strong immune response were correlated with better odds of survival.
Looking specifically at triple-negative breast cancer, a variant of breast cancer that tends to grow faster and is harder to treat, the researchers were able to identify 85 proteins associated with better odds of avoiding relapse.
In addition to reinforcing the value of proteins for studying cancer, this work could point to new targets for cancer therapeutics, as well as identify new protein biomarkers that show how cancer therapies are proceeding.
Proteomics tools for cancer
Scientists can use proteomic analyses to study the proteins that both aggravate and hinder cancer. At the moment, not every protein involved in cancer is visible to scientists. Current proteomics tools often miss low-abundance proteins, and don’t reveal a sample’s entire proteome.
With better proteomics tools, researchers may be able to confidently study more than the 4,214 proteins found in this study. That could reveal more than the 1,054 proteins used to subtype specimens here and may lead to better stratification of breast cancer types and risk. More accessible and rapid throughput technologies will help researchers efficiently perform similar studies across a wide range of cancers and other diseases.
New insights like these will hopefully be possible soon, as tools like the NautilusTM Proteome Analysis Platform are expected to give scientists unprecedented views into the full proteome of cancer cells and other biological specimens.
MORE ARTICLES