
Paleoproteomics – Unleashing the proteome of the ancient world

Tyler Ford
October 5, 2023
Paleoproteomics is the study of ancient proteins from organisms that lived long ago. In the right environments, proteins can remain largely intact for hundreds, thousands, even millions of years. By contrast, DNA molecules typically last just tens of thousands of years at most, and often far less. Proteins offer rich information about ancient creatures and cultures that other archaeological or paleontological techniques cannot access.
As novel proteomics technologies enable new proteomics applications, a deeper analysis of ancient life will become possible. For instance, proteins can be used to identify species and compare them over time, such as in comparisons between Homo sapiens and Homo neanderthalensis. Other applications of paleoproteomics include revealing what humans ate thousands of years ago, reconstructing ancient trade routes, identifying bone fragments, and more. Such efforts have already helped reconstruct the proteomes of extinct creatures. In a 2012 Journal of Proteome Research paper, a team characterized more than 100 proteins from the femur of a mammoth.
There is still much to learn from ancient proteins, and we are only beginning to truly realize the value of paleoproteomics. Today’s technologies only scratch the surface of what we can learn from ancient proteins by applying proteomics. Many proteins are still invisible to paleoproteomics techniques, meaning a vast “dark proteome” hides inside organic samples waiting to be found.
Check out this episode of the Translating Proteomics podcast to learn more about applications of proteomics
Techniques for paleoproteomics
Currently, mass spectrometry is the standard technique for analyzing ancient proteins. The proteomics technique involves running samples through a mass spectrometer, which separates components by their mass/charge ratios. Comparing these ratios to a database of known molecules enables researchers to infer protein identity.
Mass spectrometry has been crucial to the advent of paleoproteomics, enabling many exciting discoveries in the field. Some drawbacks to using mass spec techniques in paleoproteomics remain, however. Samples of ancient bones or other organic matter typically contain far fewer proteins than their modern-day counterparts. This can make it challenging to identify proteins, especially ones that were not abundant to begin with.
Greater instrument sensitivity could help find low abundance proteins, while better dynamic range would enable scientists to detect both high and low-abundance proteins at the same. Together, these improvements to proteomics tools could help power new discoveries.
Check out these animations to learn how the Nautilus Platform is designed to quantify proteins across the wide dynamic range of the proteome.
Applications of paleoproteomics
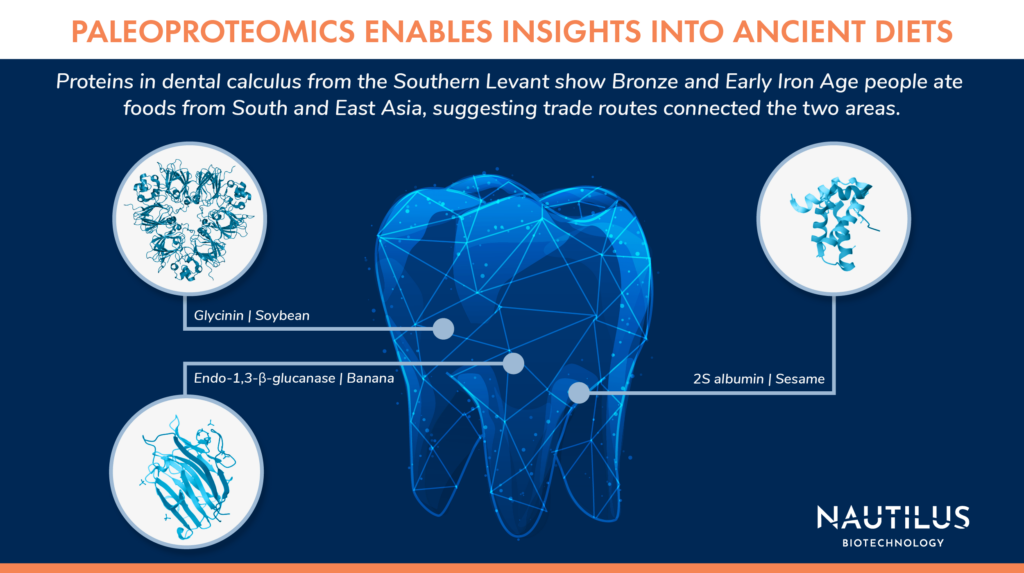
One common source of ancient proteins is in dental calculus, or plaque. Proteins from foods consumed by ancient humans, like milk and plants, as well as from bacteria inhabiting their mouths get stuck in the plaque and can be preserved for thousands of years. Studies of milk proteins in dental calculus have shown that people in Europe were practicing dairy pastoralism 5,000 years ago, and similar paleoproteomics work has revealed new evidence of trade routes between South Asia and the Near East over 3,000 years ago.
Proteins can also be detected in bones from ancient people and animals. Bones have often been broken or worn away such that just fragments remain. This can pose a challenge when researchers attempt to use standard methods to identify the species of the sample. A new project called FINDER is using proteomic analyses of collagen proteins from ancient bones to efficiently identify species even from tiny samples. Their findings could show, for example, which animals ancient humans ate, or reveal that two human lineages – like humans and Denisovans – coexisted at a site.
The future of paleoproteomics
A recent review of paleoproteomics notes that mass spectrometry often enables identification of only a small percentage of the spectra it generates. Much of the proteomic information hidden in ancient samples is yet to be revealed.
With next-generation proteomics tools capable of conducting more sensitive proteomic analysis, this ancient dark proteome could come into the light. The future of paleoproteomics will hold new and exciting insights into our past. As with so many aspects of the proteomics revolution, we are only just getting started.
Check out this episode of the Translating Proteomics podcast to learn more about why we’re poised for a proteomics breakthrough:
MORE ARTICLES
