
Applications of proteomics in neuroscience – Uncovering protein networks in Alzheimer’s

Tyler Ford
September 12, 2023
As researchers hunt for potential Alzheimer’s treatments, multiomics approaches are revealing new avenues of exploration that could lead to breakthroughs. This includes linking the proteome to the genome and other –omes to better understand the mechanisms of disease.
Alzheimer’s disease is a degenerative neurological condition that affects nearly 6 million Americans. That number could grow to 14 million by 2050. The disease is associated with buildups of amyloid beta protein and tau protein in the brain and a corresponding loss of memory and brain function.
Exactly how these two proteins work to degrade neurological function, and what other mechanisms might be involved, aren’t fully clear. Indeed, recent failures of treatments targeting amyloid beta have called into question the role of amyloid beta in Alzheimer’s and indicate that it may not be as fundamental to disease progression as was thought.
Discovery proteomics uncovers protein modules associated with Alzheimer’s
Work linking proteomics to genomics could help get us closer to an understanding of Alzheimer’s disease pathology. In a recent proteomic analysis, researchers associated protein networks with Alzheimer’s and compared them to RNA networks to see whether they match up. In this and similar studies, it is important to look to proteins and not just RNA because many proteins are modified after they are made. This means RNA expression alone does not reveal the abundance or form of fully functional proteins in many cases.
To get around that limitation and better understand how Alzheimer’s works at a fundamental level, Johnson, Carter, and Dammer, et al turned to a technique called discovery proteomics. The team looked at post mortem brain proteomes of people with Alzheimer’s, and compared them to post mortem brain proteomes of people without the disease.
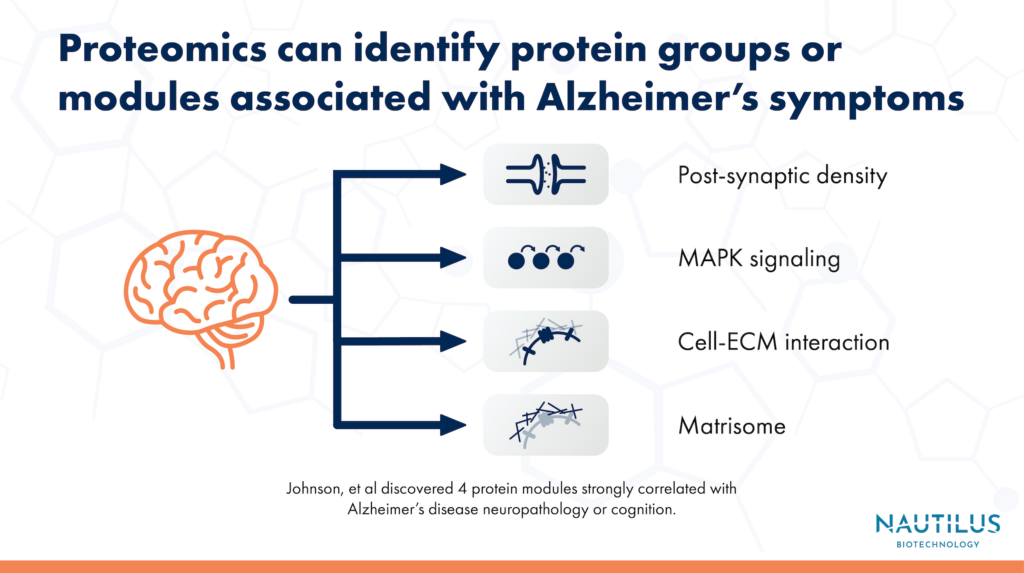
Using tandem mass tag mass spectrometry (TMT-MS), the researchers conducted a proteomic analysis of over 1,000 brain tissues from the Accelerating Medicines Partnership for Alzheimer’s Disease (AMP-AD) consortium. They identified thousands of proteins, which they grouped into 44 different co-expression modules, or groups of proteins that tend to be produced together. Of the 44 protein co-expression modules, the researchers identified 4 that were most strongly correlated with symptoms of Alzheimer’s disease.
The researchers also performed a transcriptomic analysis on post-mortem brain tissue from people with and without Alzheimer’s to determine if they could find RNA modules similar to the protein modules. This multiomics approach found that almost half of the protein modules didn’t correlate with RNA modules. This is a strong sign that Alzheimer’s disease is caused in part by post-translational modifications. The finding also underscores the value of studying the proteome to better understand Alzheimer’s.
Two of the protein modules that didn’t have any analog in the RNA modules were also ones that had the strongest connection to Alzheimer’s symptoms. One of the two modules was associated with the MAPK signaling pathway and metabolism. The other was associated with the matrisome, or the collection of proteins that make up the extracellular matrix. Proteins from both modules could be valuable targets for future research into Alzheimer’s treatments.
Furthering the multomic nature of their analysis, the researchers go on to find associations between genomic variants and the identified protein modules. In a sort of validation of this strategy, a single nucleotide polymorphism (SNP) in a gene known to impact Alzheimer’s disease risk (APOE) strongly impacted the matrisome protein module. A variety of other modules were impacted by other SNPs and, often, the SNPs were found outside of genes encoding proteins in the modules. This indicates that the SNPs impact the modules through trans effects. These findings both highlight the complexity of multiomic interactions affecting Alzheimer’s disease and point to potential targets for therapeutic intervention.
Next-generation tools for in-depth proteomic analysis
One key takeaway from this research is that extending disease studies to the proteome and covering more proteins can help uncover functional networks that were previously hidden. We’re designing the Nautilus Proteome Analysis Platform to cover nearly the entire proteome and make similar studies more accessible to more researchers. The platform’s single-molecule sensitivity and wide dynamic are designed to reveal high and low abundance proteins for unprecedented proteome coverage.
Proteomics technologies that identify all the proteins in a sample will hopefully accelerate more fantastic work like the research highlighted here. This acceleration could advance efforts to reveal the biological underpinnings of diseases like Alzheimer’s. Such studies may lead to new protein biomarkers that track disease progression and even identify targets for treatments. With next-generation proteomics, new Alzheimer’s diagnostics and treatments could be in sight.
Check out our round up of proteomics and neuroscience posts.
Watch this animation to learn how next-generation proteomics technologies can fuel neuroscience.
MORE ARTICLES

