At a casual glance, genomics and proteomics are just two different approaches to asking the same question: What’s going on inside of a cell, at the molecular level, that makes it tick? Either approach could be used to ask basic questions like why any two people are unique, or what’s causing a disease. But in reality, genomics and proteomics are complementary approaches. Combining the two will give researchers an unprecedented view of the inner workings of life, from the blueprint that encodes the architecture of our bodies to the molecules that carry out the fundamental processes underpinning life as we know it.
What is proteomics?
The definition of proteomics is the study of all the proteins – the molecular machines that carry out most biological processes – in a biological sample. Collectively, these proteins form the proteome, whether it’s in a cell, a tissue, a whole organism, or even a species.
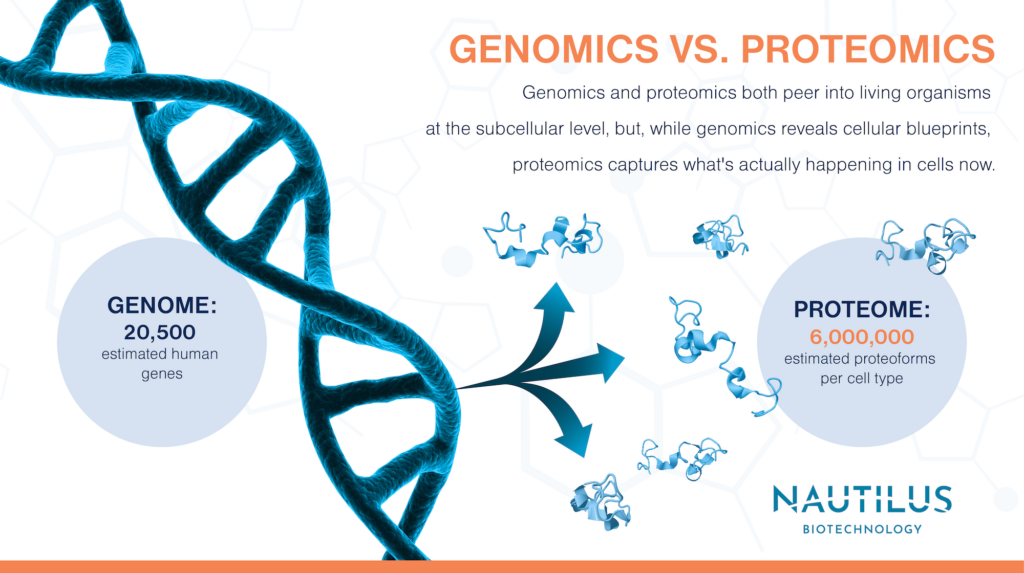
In this way, proteomics is indeed analogous to genomics. Genomics is the study of the collection of all the genetic material – the genome – in a biological sample. But if these two definitions seem the same, look closer. Genomics and proteomics both peer into living organisms at the subcellular level, but, while genomics reveals cellular blueprints, proteomics captures what’s actually happening in cells now.
Check out the Translating Proteomics podcast to learn why we’re poised for a proteomics revolution
How the genome relates to the proteome
Within an individual organism, the genome is generally the same across all the cells. Some exceptions might arise thanks to naturally occurring mutations or esoteric biological processes like immune cell development, but in general, the genes in an eyeball are the same as the genes in a pinky toe. The proteome, on the other hand, is another matter: Dynamic differences in protein composition are the reason an eyeball cell functions differently from a toe.
The proteome can be so different across cells thanks to a variety of fascinating biological processes including:
- Epigenetic modification – Factors that determine how much, if at all, a gene is expressed.
- Alternative splicing – A cellular process resulting in multiple ways to read a gene.
- Post-translational modifications PTMs – Alterations such as the addition of small molecules to translated proteins that can change protein function.
The size of the genome vs. proteome
Any given individual gene – a segment of DNA that encodes a functional RNA (many of which are translated into proteins) – might be hundreds or thousands of base pairs long. In humans, scientists estimate there are around 20,000 of these coding genes. But this doesn’t mean that there are 20,000 proteins. Proteins come in many different forms called proteoforms, and there are many of them.
How many human proteoforms are there? Splice variants alone take the number of possible proteins from ~20,000 up to ~70,000 proteoforms. Then possible combinations of PTMs could add as many as 100,000 more proteoform variations per protein. With more and more potential variations, the number of possible proteoforms becomes, as researchers put it, astronomically large. But that’s a theoretical maximum, not necessarily what a biological sample actually contains. A better guess is six million.
Much more research is needed to take this number from a napkin-math estimate to an actual measurement of nature. Studies so far have been hampered by the technology researchers use to study the proteome. Traditional proteomics analysis methods like mass spectroscopy can only measure a fraction of the proteins in a cell at a time. But with next-generation proteomics technologies like the Nautilus platform, this may soon change. The PrISM framework at the heart of the Nautilus proteomics platform is designed to identify and quantify over 95% of the proteins in a biological sample.
Genomics vs. proteomics applications
The study of genes and proteins can achieve different purposes, both in research and clinical applications. Which one researchers should use depends on what they’d like to learn, but most often, the two approaches complement each other.
There are some instances where a gene’s effects are well understood, and genomic approaches are well suited to answering questions. For instance, a certain mutation may cause a disease, or could confer resistance to a disease, in which case genetic testing would be an appropriate approach. Any time researchers are looking for the root of a biological process, it can make sense to trace it all the way back to the genes.
But as we’ve outlined above, genes aren’t always expressed the same way. They determine what proteins cells can possibly make, but the proteins themselves are doing the heavy lifting. Having a comprehensive genome sequence in-hand for an organism does not reveal the full story what’s happening in a cell or sample of interest now. When studying the mechanisms of on-going cellular processes, it’s far more useful to also look at the proteome to see what proteins are present, in what abundances, and how they’re interacting with one another to carry out cellular functions.
When developing therapeutics, researchers really need to understand what’s happening at the protein level. That’s why proteomic analysis, rather than genomic analysis, can be a more direct route to drug development. Knowing what proteins are over- or under- represented in a disease state can help researchers identify biomarkers for diagnosis as well as a specific protein targets for drugs. For example, proteomics can help in the development of precision medicine against cancer,
Another promising avenue for proteomics in disease research is learning how proteoforms impact disease. For instance, Alzheimer’s disease is associated with changes to a protein called “tau.” This is an incredibly complex protein known for having many post-translational modifications. Tau studies are a prime example of how proteomics, rather than genomics, is required to understand the complex relationships between health and disease.
Find more applications of proteomics.
Watch this animation to learn how proteomics can drive cancer research:
Combining genomics and proteomics for a better future
As we enter an era where scientists leverage biotechnologies to solve a wide array of existential crises, looking at the genome or proteome in isolation won’t cut it. Researchers must leverage both of these powerful sources of biological insight to drive the development of novel solutions that create a better future for everyone. New technologies like the Nautilus Proteome Analysis Platform are designed to help us achieve this better future.
Want to learn more? Check out Nautilus’ recent white paper series on “The proteomics revolution” for more on the promise of proteomics.
MORE ARTICLES


